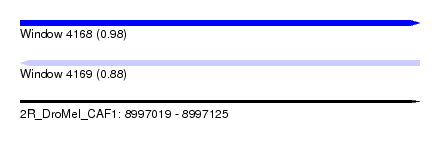
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,997,019 – 8,997,125 |
| Length | 106 |
| Max. P | 0.984472 |

| Location | 8,997,019 – 8,997,125 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 72.09 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -16.08 |
| Energy contribution | -15.13 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.984472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
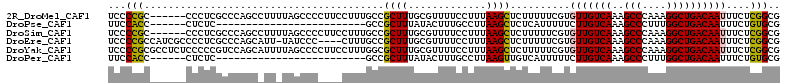
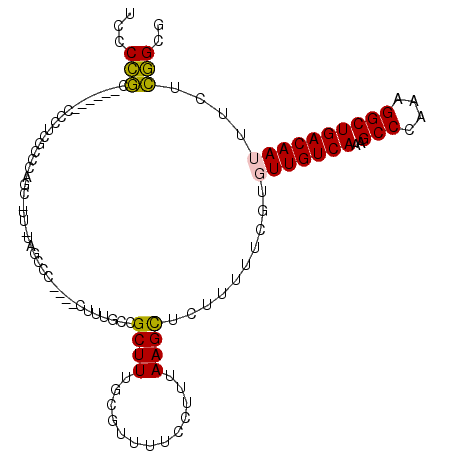
>2R_DroMel_CAF1 8997019 106 + 20766785 CGCCGAGAAAUUGUCAGCCUUUGGGCUUUGACAACACGAAAAAGAGCUUAAAGGAAAACGCAAAGCGGCAAAGGAAGGGGCUAAAAGGCUGGGCGAGGG------GCGGGGA ((((.............((((((((((((.............))))))))))))....(((..((((((..........))).....)))..)))...)------))).... ( -30.42) >DroPse_CAF1 25109 81 + 1 CGCACAGAAAUUGUCAGCCAAAGGGCUUUGACAAGAAAAAUGAGAGCUUAAGGCAAAGUAUAAAGCGGC-------------------------GAGAG------GGUGGAA ..(((.....(((((.(((...(((((((.(.........).)))))))..)))...((.....)))))-------------------------))...------.)))... ( -17.50) >DroSim_CAF1 27623 106 + 1 CGCCGAGAAAUUGUCAGCCUUUGGGCUUUGACAACACGAAAAAGAGCUUAAAGGAAAACGCAAAGCGGCAAAGGAAGGGGCUAAAAGGCUGGGCGAGGG------GCGGGGA ((((.............((((((((((((.............))))))))))))....(((..((((((..........))).....)))..)))...)------))).... ( -30.42) >DroEre_CAF1 27880 107 + 1 CGCCGAGAAAUUGUCAGCCUUUGGGCUUUGACAACACGAAAAAGAGCUUAAAGGAAAACGCAAAGCGGCAAAG----GGGAUA-AAUGCUGGGCGAGGGGCGAUGGCGGGGA (.(((....((((((..((((((((((((.............))))))))))))....(((..((((......----......-..))))..)))...))))))..))).). ( -34.36) >DroYak_CAF1 28374 112 + 1 CGCCGAGAAAUUGUCAGCCUUUGGGCUUUGACAACACGAAAAAGAGCUUAAAGGAAAACGCAAAGCGCCAAAGGAAGGGGCUAAAAUGCUGGACGGGGGAGAGGCGCGGGGA ((((......(((((..((((((((((((.............)))))))))))).....(((.(((.((.......)).)))....)))..)))))......))))...... ( -34.42) >DroPer_CAF1 24393 81 + 1 CGCACAGAAAUUGUCAGCCAAAGGGCUUUGACAAGAAAAAUGACAACUUAAGGCAAAGUAUAAAGCGGC-------------------------GAGAG------GGUGGAA ..(((.....(((((((((....)))..))))))............(((...((...((.....)).))-------------------------...))------))))... ( -15.60) >consensus CGCCGAGAAAUUGUCAGCCUUUGGGCUUUGACAACACGAAAAAGAGCUUAAAGGAAAACGCAAAGCGGCAAAG____GGGCUA_AA_GCUGGGCGAGGG______GCGGGGA ..(((.....(((((((((....)))..))))))...........((((...(.....)...))))........................................)))... (-16.08 = -15.13 + -0.94)

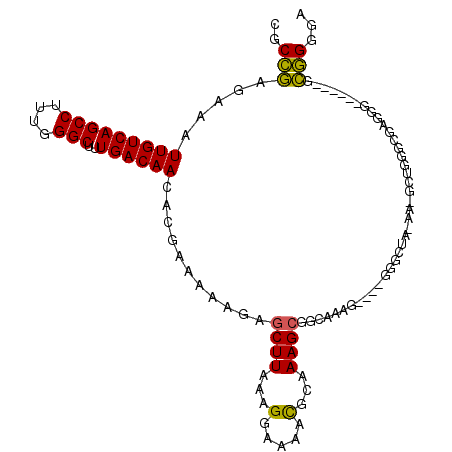
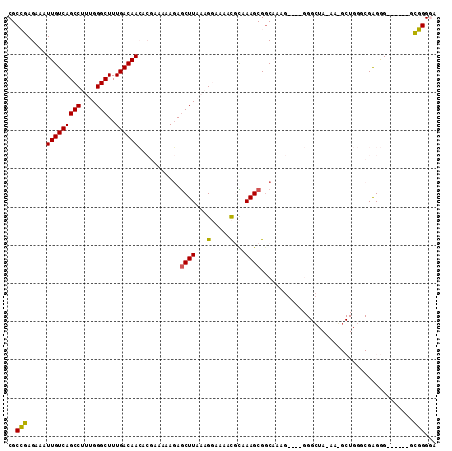
| Location | 8,997,019 – 8,997,125 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 72.09 |
| Mean single sequence MFE | -18.85 |
| Consensus MFE | -13.19 |
| Energy contribution | -12.50 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8997019 106 - 20766785 UCCCCGC------CCCUCGCCCAGCCUUUUAGCCCCUUCCUUUGCCGCUUUGCGUUUUCCUUUAAGCUCUUUUUCGUGUUGUCAAAGCCCAAAGGCUGACAAUUUCUCGGCG ....(((------(...(((..(((.....((.......)).....)))..))).......................(((((((..(((....)))))))))).....)))) ( -19.90) >DroPse_CAF1 25109 81 - 1 UUCCACC------CUCUC-------------------------GCCGCUUUAUACUUUGCCUUAAGCUCUCAUUUUUCUUGUCAAAGCCCUUUGGCUGACAAUUUCUGUGCG .......------....(-------------------------(((((((.............))))...........((((((..(((....))))))))).....).))) ( -12.92) >DroSim_CAF1 27623 106 - 1 UCCCCGC------CCCUCGCCCAGCCUUUUAGCCCCUUCCUUUGCCGCUUUGCGUUUUCCUUUAAGCUCUUUUUCGUGUUGUCAAAGCCCAAAGGCUGACAAUUUCUCGGCG ....(((------(...(((..(((.....((.......)).....)))..))).......................(((((((..(((....)))))))))).....)))) ( -19.90) >DroEre_CAF1 27880 107 - 1 UCCCCGCCAUCGCCCCUCGCCCAGCAUU-UAUCCC----CUUUGCCGCUUUGCGUUUUCCUUUAAGCUCUUUUUCGUGUUGUCAAAGCCCAAAGGCUGACAAUUUCUCGGCG ....((((.........(((..(((...-......----.......)))..))).......................(((((((..(((....)))))))))).....)))) ( -20.89) >DroYak_CAF1 28374 112 - 1 UCCCCGCGCCUCUCCCCCGUCCAGCAUUUUAGCCCCUUCCUUUGGCGCUUUGCGUUUUCCUUUAAGCUCUUUUUCGUGUUGUCAAAGCCCAAAGGCUGACAAUUUCUCGGCG ......((((.............(((....(((.((.......)).))).)))((((......))))..........(((((((..(((....)))))))))).....)))) ( -24.90) >DroPer_CAF1 24393 81 - 1 UUCCACC------CUCUC-------------------------GCCGCUUUAUACUUUGCCUUAAGUUGUCAUUUUUCUUGUCAAAGCCCUUUGGCUGACAAUUUCUGUGCG .......------.....-------------------------...............(((..(((((((((........((((((....)))))))))))))))..).)). ( -14.60) >consensus UCCCCGC______CCCUCGCCCAGC_UU_UAGCCC____CUUUGCCGCUUUGCGUUUUCCUUUAAGCUCUUUUUCGUGUUGUCAAAGCCCAAAGGCUGACAAUUUCUCGGCG ...(((........................................((((.............))))..........(((((((..(((....))))))))))....))).. (-13.19 = -12.50 + -0.69)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:18 2006