
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,996,265 – 8,996,399 |
| Length | 134 |
| Max. P | 0.942071 |

| Location | 8,996,265 – 8,996,359 |
|---|---|
| Length | 94 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 90.21 |
| Mean single sequence MFE | -20.66 |
| Consensus MFE | -18.06 |
| Energy contribution | -17.75 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
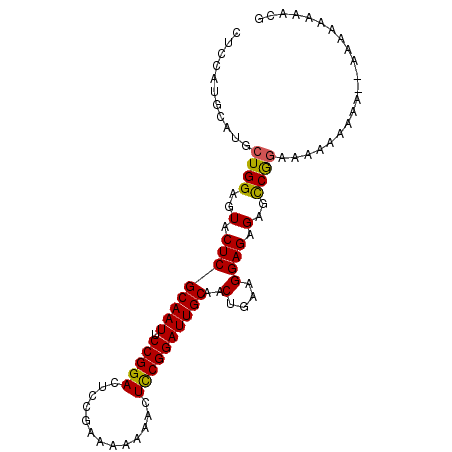
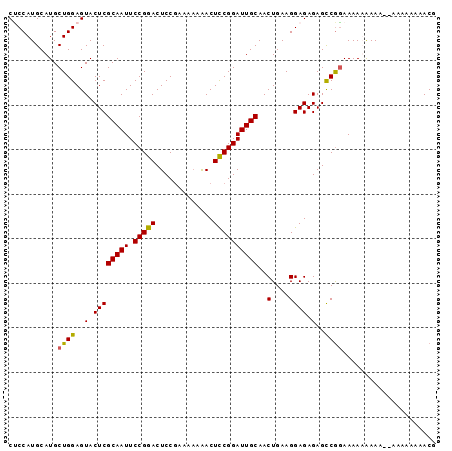
>2R_DroMel_CAF1 8996265 94 + 20766785 CCCCAUGCAUGCUGGAGUACUCGCAAUUCCGGACUCCGAAAA-AACUUCGGAUUGCAACUGAAGGAGAGAGCCGGAAAAAAAAA-AUAAAAAAACG ..((.((((..(((((((.......)))))))..((((((..-...)))))).))))......((......)))).........-........... ( -20.70) >DroSec_CAF1 26598 94 + 1 CUCCAUGCAUGCUGGAGUACUCGCAAUUCCGGACUCCGAAAAAGACUCCGGAUUGCAACUGAAGGAGAGAGCCGGAAAAUAAAA--AAAAUAAACG (((((.(....))))))..((((((((.(((((.((.......)).))))))))))..((.....)).))).............--.......... ( -22.40) >DroSim_CAF1 26847 96 + 1 CUCCAUGCAUGCUGGAGUACUCGCAAUUCCGGACUCCGAAAAAAACUCCGGAUUGCAACUGAAGGAGAGAGCCGGAAAAUAAAAAAAAAAUAAACG (((((.(....)))))).........((((((.(((.........((((..............)))).)))))))))................... ( -20.24) >DroYak_CAF1 27619 91 + 1 CGCCAUGCAUGCUGGAGUACUCGCAAUUCCGGACUCCAAA--AAACUCCGGAUUGCAACUGAAGGAGAGAGUCACAAAAAGAA---AAAAAAAACG ..(((.(....)))).(((((((((((.(((((.......--....))))))))))..((.....)).)))).))........---.......... ( -19.30) >consensus CUCCAUGCAUGCUGGAGUACUCGCAAUUCCGGACUCCGAAAAAAACUCCGGAUUGCAACUGAAGGAGAGAGCCGGAAAAAAAAA__AAAAAAAACG ...........((((..(.((((((((.(((((.............))))))))))..(....)))).)..))))..................... (-18.06 = -17.75 + -0.31)

| Location | 8,996,292 – 8,996,399 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 91.69 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -21.60 |
| Energy contribution | -21.23 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
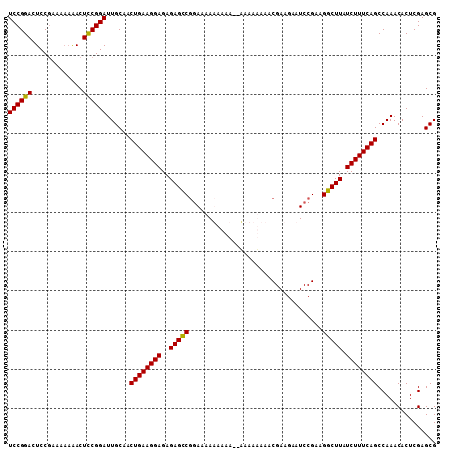
>2R_DroMel_CAF1 8996292 107 + 20766785 UCCGGACUCCGAAAA-AACUUCGGAUUGCAACUGAAGGAGAGAGCCGGAAAAAAAAA-AUAAAAAAACGAAGAAUCCGAAGGCUUAUCUUUCAGCCAAACACUCGAGCG ...((..((((((..-...))))))......((((((((..((((((((........-................)))...))))).))))))))))............. ( -24.96) >DroSec_CAF1 26625 107 + 1 UCCGGACUCCGAAAAAGACUCCGGAUUGCAACUGAAGGAGAGAGCCGGAAAAUAAAA--AAAAUAAACGAAGAAUCCGAAGGCUUAUCUUUCAGCCAAACACUCGAGCG ((((((.((.......)).))))))......((((((((..((((((((........--...............)))...))))).))))))))............... ( -26.20) >DroSim_CAF1 26874 109 + 1 UCCGGACUCCGAAAAAAACUCCGGAUUGCAACUGAAGGAGAGAGCCGGAAAAUAAAAAAAAAAUAAACGAAGAAUCCGAAGGCUUAUCUUUCAGCCAAACACUCGAGCG ((((((.............))))))......((((((((..((((((((.........................)))...))))).))))))))............... ( -23.73) >DroYak_CAF1 27646 104 + 1 UCCGGACUCCAAA--AAACUCCGGAUUGCAACUGAAGGAGAGAGUCACAAAAAGAA---AAAAAAAACGAAGGAUCCGAAGGCUUAUCUUUCAGCCAAACACUCGAGCG ((((((.......--....))))))......((((((((..(((((.(.....)..---........(....).......))))).))))))))............... ( -20.80) >consensus UCCGGACUCCGAAAAAAACUCCGGAUUGCAACUGAAGGAGAGAGCCGGAAAAAAAAA__AAAAAAAACGAAGAAUCCGAAGGCUUAUCUUUCAGCCAAACACUCGAGCG ((((((.............))))))......((((((((..(((((..................................))))).))))))))............... (-21.60 = -21.23 + -0.38)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:16 2006