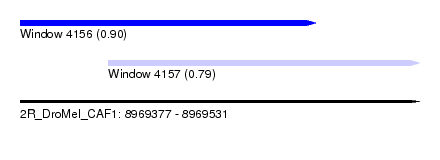
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,969,377 – 8,969,531 |
| Length | 154 |
| Max. P | 0.900891 |

| Location | 8,969,377 – 8,969,491 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.70 |
| Mean single sequence MFE | -16.53 |
| Consensus MFE | -10.70 |
| Energy contribution | -11.70 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.900891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8969377 114 + 20766785 UGAAAGACUAUUCAAAUCCAUAUCAUUUUUUAUAAAAUCGAAUUUACCAUCGUUAGGUAGAAUGUGUGUAUAAUACAUACAUUUUACUUUACACGAUAUUAACUUCCACUCUAC ((((......)))).........................(((.(((..(((((.((((((((((((((((...))))))))))))))))...)))))..))).)))........ ( -23.90) >DroSec_CAF1 16671 110 + 1 GA-AAGACUAUUCAAAUCCAUAUCAUUUUUUAUAAAAUCGAACUUACCA---UAAAGUAGAAUAAGUGUAUAAUACAUACAUUUUACUUUACACGAUAUUAACUUCCACUCUAC ..-................((((((((((....)))))...........---(((((((((((..(((((...)))))..)))))))))))...)))))............... ( -13.00) >DroYak_CAF1 13540 108 + 1 -GAAUGACUAUUCAAAUCCAUAUCAACUUUUAUAAAAACUUUUUUACUUUC-----GUAGAAAGUGUGUAUAAUACGUAUAUUUUACCUUACACGAUAUAAACUUCCACUCCAC -((((....))))......(((((.............(((((((.((....-----)))))))))(((((......(((.....)))..))))))))))............... ( -12.70) >consensus _GAAAGACUAUUCAAAUCCAUAUCAUUUUUUAUAAAAUCGAAUUUACCAUC_U_A_GUAGAAUGUGUGUAUAAUACAUACAUUUUACUUUACACGAUAUUAACUUCCACUCUAC ...................(((((........((((......))))..........((((((((((((((...)))))))))))))).......)))))............... (-10.70 = -11.70 + 1.00)

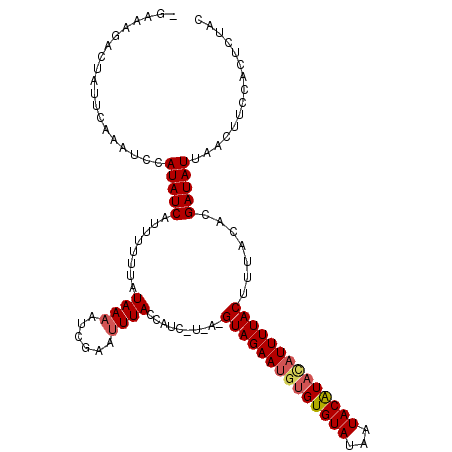
| Location | 8,969,411 – 8,969,531 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.55 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -14.26 |
| Energy contribution | -15.06 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8969411 120 + 20766785 AAAUCGAAUUUACCAUCGUUAGGUAGAAUGUGUGUAUAAUACAUACAUUUUACUUUACACGAUAUUAACUUCCACUCUACAUUACCUGGUGGCGCCAUCUAGGAGUCCCGGUUAGACAAU .....(((.(((..(((((.((((((((((((((((...))))))))))))))))...)))))..))).)))...((((....(((.((.(((.((.....)).)))))))))))).... ( -34.80) >DroSec_CAF1 16704 116 + 1 AAAUCGAACUUACCA---UAAAGUAGAAUAAGUGUAUAAUACAUACAUUUUACUUUACACGAUAUUAACUUCCACUCUACAUUCCCCA-UGGCGCCAUCUAGGAGUGCCAGUUAGACAAU ...((.((((.....---(((((((((((..(((((...)))))..)))))))))))......................((((((..(-((....)))...))))))..)))).)).... ( -21.20) >DroSim_CAF1 13895 120 + 1 AUAUCGAAUUUACCAUCGUAAAGUAGAAUGUGUGUAUAUUACGUACAUUUUACCUUACACAAUAUUAACUUCCACUCUACAUUCCCCAGUGGCGCCAUCUAGGAGUGCCAGUUAGACAAA .................((((.((((((((((((((...)))))))))))))).))))......((((((..((((((..........(((....)))...))))))..))))))..... ( -25.72) >DroEre_CAF1 13723 114 + 1 AAAACUUUUUUUCU-UC-----GUAGAAAAUGAGUAUUAUACGUAUAUUUUACUUUACACAAGAUUAACAUCCACUCUACAUUCCCAGGUGGCGCCAUCUAGUCGUGCCAGUUUAAUAAU ...((((.((((((-..-----..)))))).))))..........((((..(((.(((((.((((......((((.((........))))))....)))).)).)))..)))..)))).. ( -18.10) >DroYak_CAF1 13573 115 + 1 AAAACUUUUUUACUUUC-----GUAGAAAGUGUGUAUAAUACGUAUAUUUUACCUUACACGAUAUAAACUUCCACUCCACAUUCCCAGGUGGCGCCAUCUAGCCGUGGCACUUAACCAAU ...(((((((.((....-----)))))))))(((((......(((.....)))..)))))...........................(((((.(((((......))))).))..)))... ( -19.20) >consensus AAAUCGAAUUUACCAUC_U_A_GUAGAAUGUGUGUAUAAUACGUACAUUUUACUUUACACGAUAUUAACUUCCACUCUACAUUCCCAGGUGGCGCCAUCUAGGAGUGCCAGUUAGACAAU ......................((((((((((((((...))))))))))))))....................................((((((.........)))))).......... (-14.26 = -15.06 + 0.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:32:06 2006