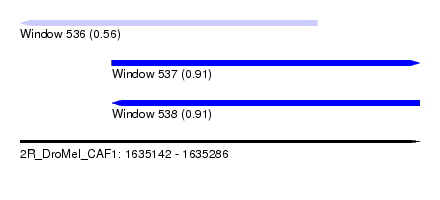
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,635,142 – 1,635,286 |
| Length | 144 |
| Max. P | 0.911634 |

| Location | 1,635,142 – 1,635,249 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.22 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -20.67 |
| Energy contribution | -20.80 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1635142 107 - 20766785 -----AGCUCCGUCAGCAGAUCUGCUUGGGAAACAAAGCAGCUGAAGGAAUACAGAGCUAAAGAAGCUUUUAAUCAAAAUGUAGAUUCCAU-------GUUAUAGCCAACUAAUUGAAA -----.(((((.(((((.....(((((.(....).)))))))))).)))....((((((.....)))))).....................-------......))............. ( -23.20) >DroSec_CAF1 20946 107 - 1 -----AGCUCCGUCAACAGAUCUGCUUGGGAAACGCAGCAGCUGAAGGAAUACAGAGCUGAAGAAGCUUUUAAUGAAAAUGUAGGUUCCAU-------GUUAUAGCCAACUAAUUGAAA -----.(((....((.(((..(((((.((....).)))))))))..(((((..((((((.....))))))..((....))....))))).)-------)....)))............. ( -22.60) >DroSim_CAF1 19378 107 - 1 -----AGCUCCGUCAACAGAUCUGCUUGGGAAACGCAGCAGCUGAAGGAAUACAGAGCUGAAGAAGCUUUUAAUGAAAAUGUAGGUUCCAU-------GUUAUAGCAAACUAAUUGAAA -----.(((....((.(((..(((((.((....).)))))))))..(((((..((((((.....))))))..((....))....))))).)-------)....)))............. ( -23.50) >DroEre_CAF1 23461 109 - 1 UACAUAGCUCCGUCAACAGAUCUGCUUGGGAAACUAAGCAGCUGAAGGAAUACAGAGCUGAAGAUGCUUUUAAUGAAAAUGUAGGGUCCAAUAACUUGGCUAUAACG----------AA ...((((((..((...(((..((((((((....)))))))))))..(((.((((((((.......)))))..((....)))))...)))....))..))))))....----------.. ( -31.30) >consensus _____AGCUCCGUCAACAGAUCUGCUUGGGAAACGAAGCAGCUGAAGGAAUACAGAGCUGAAGAAGCUUUUAAUGAAAAUGUAGGUUCCAU_______GUUAUAGCCAACUAAUUGAAA .....(((((..((..(((..((((((.(....).)))))))))...)).....)))))...(((.((....((....))..)).)))............................... (-20.67 = -20.80 + 0.13)

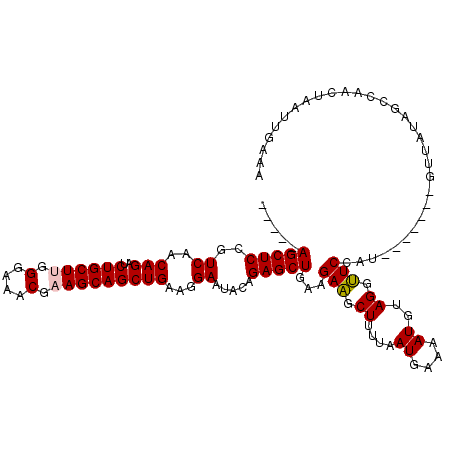
| Location | 1,635,175 – 1,635,286 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 91.30 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -20.95 |
| Energy contribution | -21.57 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.05 |
| SVM RNA-class probability | 0.906040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1635175 111 + 20766785 UUUUGAUUAAAAGCUUCUUUAGCUCUGUAUUCCUUCAGCUGCUUUGUUUCCCAAGCAGAUCUGCUGACGGAGCU--------AAAUAUCCCAGAGUUUCCUUUUAAUAAUUCACAUCUU .....((((((((.(((((((((((((.......(((((.(.((((((.....)))))).).))))))))))))--------)).......))))....))))))))............ ( -25.12) >DroSec_CAF1 20979 111 + 1 UUUUCAUUAAAAGCUUCUUCAGCUCUGUAUUCCUUCAGCUGCUGCGUUUCCCAAGCAGAUCUGUUGACGGAGCU--------AGAUAUCCCAGAGUCUCCUUUUAAUAAUUCACAUGUU .....((((((((.......((((((((.......(((...((((.........))))..)))...))))))))--------((((........)))).))))))))............ ( -24.50) >DroSim_CAF1 19411 111 + 1 UUUUCAUUAAAAGCUUCUUCAGCUCUGUAUUCCUUCAGCUGCUGCGUUUCCCAAGCAGAUCUGUUGACGGAGCU--------AGAUAUCCCAGAGUCUCCUUUCAAUAAUUCACAUGUU ...........((((((.(((((.(((........)))(((((..........)))))....))))).))))))--------((((........))))..................... ( -21.10) >DroEre_CAF1 23491 119 + 1 UUUUCAUUAAAAGCAUCUUCAGCUCUGUAUUCCUUCAGCUGCUUAGUUUCCCAAGCAGAUCUGUUGACGGAGCUAUGUAGAUAGAUAUCCCAGAGUCUCCUUUUAUUAAUUCACAUGUU .......((((((.((((..((((((((.......(((((((((........))))))..)))...))))))))....))))((((........)))).)))))).............. ( -24.50) >consensus UUUUCAUUAAAAGCUUCUUCAGCUCUGUAUUCCUUCAGCUGCUGCGUUUCCCAAGCAGAUCUGUUGACGGAGCU________AGAUAUCCCAGAGUCUCCUUUUAAUAAUUCACAUGUU .....((((((((.......((((((((.......(((((((((........))))))..)))...))))))))........((((........)))).))))))))............ (-20.95 = -21.57 + 0.62)

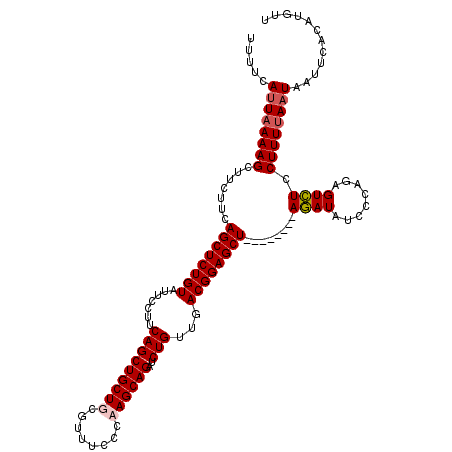
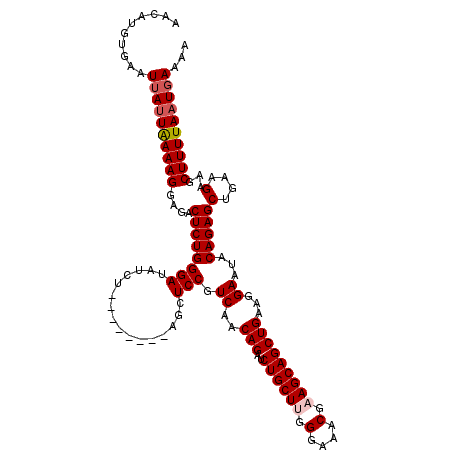
| Location | 1,635,175 – 1,635,286 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 91.30 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -23.15 |
| Energy contribution | -23.96 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1635175 111 - 20766785 AAGAUGUGAAUUAUUAAAAGGAAACUCUGGGAUAUUU--------AGCUCCGUCAGCAGAUCUGCUUGGGAAACAAAGCAGCUGAAGGAAUACAGAGCUAAAGAAGCUUUUAAUCAAAA ..(((.((((.((((...((....))....))))(((--------(((((..((..(((..((((((.(....).)))))))))...)).....))))))))......))))))).... ( -27.70) >DroSec_CAF1 20979 111 - 1 AACAUGUGAAUUAUUAAAAGGAGACUCUGGGAUAUCU--------AGCUCCGUCAACAGAUCUGCUUGGGAAACGCAGCAGCUGAAGGAAUACAGAGCUGAAGAAGCUUUUAAUGAAAA ..........((((((((((....((((((((.....--------...))).((..(((..(((((.((....).)))))))))...))...)))))((.....))))))))))))... ( -25.70) >DroSim_CAF1 19411 111 - 1 AACAUGUGAAUUAUUGAAAGGAGACUCUGGGAUAUCU--------AGCUCCGUCAACAGAUCUGCUUGGGAAACGCAGCAGCUGAAGGAAUACAGAGCUGAAGAAGCUUUUAAUGAAAA ..........((((((((((....((((((((.....--------...))).((..(((..(((((.((....).)))))))))...))...)))))((.....))))))))))))... ( -25.80) >DroEre_CAF1 23491 119 - 1 AACAUGUGAAUUAAUAAAAGGAGACUCUGGGAUAUCUAUCUACAUAGCUCCGUCAACAGAUCUGCUUGGGAAACUAAGCAGCUGAAGGAAUACAGAGCUGAAGAUGCUUUUAAUGAAAA ..((((((...........((((.((.((((((....)))).)).)))))).((..(((..((((((((....)))))))))))...)).)))(((((.......)))))..))).... ( -31.60) >consensus AACAUGUGAAUUAUUAAAAGGAGACUCUGGGAUAUCU________AGCUCCGUCAACAGAUCUGCUUGGGAAACGAAGCAGCUGAAGGAAUACAGAGCUGAAGAAGCUUUUAAUGAAAA ..........((((((((((....((((((((................))).((..(((..((((((.(....).)))))))))...))...)))))(....)...))))))))))... (-23.15 = -23.96 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:35 2006