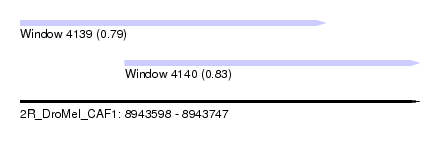
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,943,598 – 8,943,747 |
| Length | 149 |
| Max. P | 0.832838 |

| Location | 8,943,598 – 8,943,712 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.90 |
| Mean single sequence MFE | -26.70 |
| Consensus MFE | -16.74 |
| Energy contribution | -16.52 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
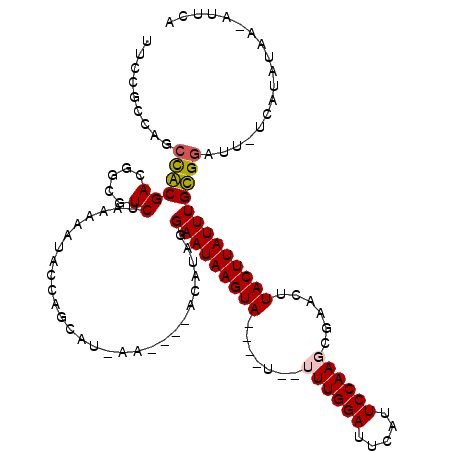
>2R_DroMel_CAF1 8943598 114 + 20766785 UUCCGCCAGCCGCGACGGCGUCUAAAAAUACCGACAU-AG----ACAUAGGAAUAAGUAGGUUUUUUUUGGAUUCAUUCCAAGCGAACUUACUUAUUUGCGGAUU-UCAUAUAAAAUUCA .(((((..((((...))))(((((............)-))----))....((((((((((((((..((((((.....)))))).)))))))))))))))))))..-.............. ( -36.40) >DroPse_CAF1 10093 108 + 1 UUCCCCCAGCUACGACGCGCUCUAAAAAUACCAGCAUCAA----ACACAGGAAUAAGUA----U--UUUGGAUUCAUUCCAAUCGUAUUUACUUAUUUGUGGAUU-UCAUAUAC-AUACA ........((......))(((...........))).....----...((.(((((((((----.--.(((((.....))))).......))))))))).))....-........-..... ( -17.00) >DroEre_CAF1 17341 111 + 1 UUCUGCCAGCCGCGACGGCGUCUAAAAAUACCACCAU-AG----ACAUAGGAAUAAGUAGGUAU--UUUGGAUUCAUUCCAAGCAAACUUACUUAUUUGCGGAUU-UCAUAUAA-AUUCA .(((((..((((...))))(((((............)-))----))....((((((((((((..--((((((.....))))))...)))))))))))))))))..-........-..... ( -32.00) >DroYak_CAF1 12374 115 + 1 UUCCGCCAGCCGCGACGGCGUCUAAAAAUACAACCGU-AAAAAUACAUGGGAAUAAGUAGGUAU--UUUGGAUUCAUUCCAAGCGAACUUACUUAUUUGCGGAUU-UCAUAUAA-AUUCA .(((((..((((...))))..............((((-........))))((((((((((((..--((((((.....))))))...)))))))))))))))))..-........-..... ( -33.20) >DroAna_CAF1 14978 102 + 1 UUCCGCCGGACACGACGGCAUCUAAAAAUACCAGC-----------AUAGGAAUAAGUA----U--UUUGGAUUCAUUCCAAGCACAUUUACUUAUUUGUGGAUUUUCAUAUAA-AUCCA ....((((.......))))................-----------..(.(((((((((----.--((((((.....))))))......))))))))).)((((((......))-)))). ( -24.60) >DroPer_CAF1 10168 108 + 1 UUCCCCCAGCUACGACGCGCUCUAAAAAUACCAGCAUCAA----ACACAGGAAUAAGUA----U--UUUGGAUUCAUUCCAAUCGUAUUUACUUAUUUGUGGAUU-UCAUAUAC-AUACA ........((......))(((...........))).....----...((.(((((((((----.--.(((((.....))))).......))))))))).))....-........-..... ( -17.00) >consensus UUCCGCCAGCCACGACGGCGUCUAAAAAUACCAGCAU_AA____ACAUAGGAAUAAGUA____U__UUUGGAUUCAUUCCAAGCGAACUUACUUAUUUGCGGAUU_UCAUAUAA_AUUCA .........((((((.....))............................(((((((((.......((((((.....))))))......))))))))))))).................. (-16.74 = -16.52 + -0.22)

| Location | 8,943,637 – 8,943,747 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.75 |
| Mean single sequence MFE | -28.34 |
| Consensus MFE | -21.18 |
| Energy contribution | -21.43 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
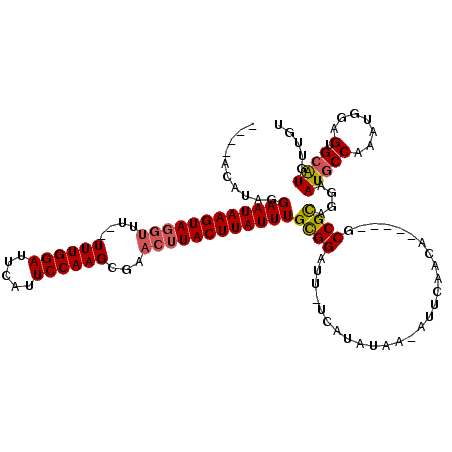
>2R_DroMel_CAF1 8943637 110 + 20766785 ----ACAUAGGAAUAAGUAGGUUUUUUUUGGAUUCAUUCCAAGCGAACUUACUUAUUUGCGGAUU-UCAUAUAAAAUUCAACA-----GCCGCAGGAAUGCCAAAUGGUGUGCAUUUUGA ----......((((((((((((((..((((((.....)))))).))))))))))))))((((...-.................-----.)))).(((((((((.....)).))))))).. ( -30.80) >DroSec_CAF1 17904 108 + 1 ----ACAUAGGAAUAAGUAGGUUUU-UUUGGAUUCAUUCCAAGCGAACUUACUUAUUUGCGGAUU-UCAUAUAA-AUUCAACA-----GCCGCAGGAAUGCCAAAUGGAGUGCAUCUUGU ----......((((((((((((((.-((((((.....)))))).))))))))))))))..(((((-(.....))-))))....-----...((((((.((((.......).))))))))) ( -31.40) >DroSim_CAF1 12181 108 + 1 ----ACAUAGGAAUAAGUAGGUUUU-UUUGGAUUCAUUCCAAGCGAACUUACUUAUUUGCGGAUU-UCAUAUAA-AUUCAACA-----GCCGCAGGAAUGCCAAAUGGAGUGCAUGUUGU ----......((((((((((((((.-((((((.....)))))).))))))))))))))..(((((-(.....))-)))).(((-----((.(((......((....))..)))..))))) ( -30.10) >DroEre_CAF1 17380 104 + 1 ----ACAUAGGAAUAAGUAGGUAU--UUUGGAUUCAUUCCAAGCAAACUUACUUAUUUGCGGAUU-UCAUAUAA-AUUCA--------GCCGCAGAAAUGCCAAAUGGAGUGCAUGAUAU ----.(((..((((((((((((..--((((((.....))))))...))))))))))))(((((((-(.....))-)))).--------...(((....)))..........))))).... ( -26.80) >DroYak_CAF1 12413 108 + 1 AAAUACAUGGGAAUAAGUAGGUAU--UUUGGAUUCAUUCCAAGCGAACUUACUUAUUUGCGGAUU-UCAUAUAA-AUUCA--------ACCGCAGGAAUGCCCAAUGGAGUGCAUGCUAU .....(((((((((((((((((..--((((((.....))))))...)))))))))))(((((...-........-.....--------.)))))......))).))).(((....))).. ( -28.43) >DroAna_CAF1 15013 107 + 1 ------AUAGGAAUAAGUA----U--UUUGGAUUCAUUCCAAGCACAUUUACUUAUUUGUGGAUUUUCAUAUAA-AUCCAAAACAAAUUCCGUUGAAAUGCCUAAUAAAGUGAGUUCACA ------.((((((((((((----.--((((((.....))))))......))))))))..(((((((......))-)))))....................)))).....(((....))). ( -22.50) >consensus ____ACAUAGGAAUAAGUAGGUUU__UUUGGAUUCAUUCCAAGCGAACUUACUUAUUUGCGGAUU_UCAUAUAA_AUUCAACA_____GCCGCAGGAAUGCCAAAUGGAGUGCAUGUUGU ..........((((((((((((....((((((.....))))))...))))))))))))((((...........................))))....(((((.......).))))..... (-21.18 = -21.43 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:50 2006