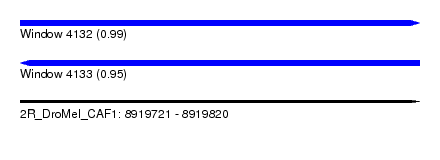
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,919,721 – 8,919,820 |
| Length | 99 |
| Max. P | 0.994030 |

| Location | 8,919,721 – 8,919,820 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
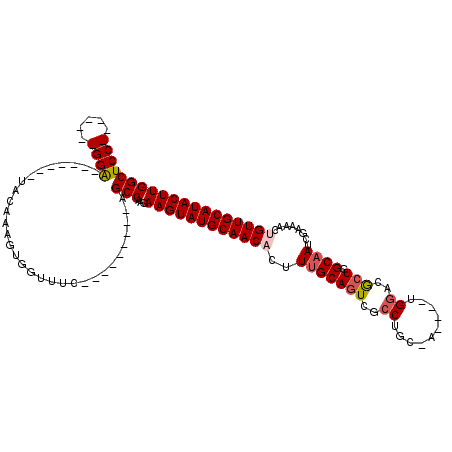
| Reading direction | forward |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -35.32 |
| Consensus MFE | -24.64 |
| Energy contribution | -25.60 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.994030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8919721 99 + 20766785 GCCAAGUAUGCAACAGUUUUCGAUUGCCAGCGUCCA----U-GCAGGCGACUGCAAAGUGUUGCAUACUUCUUGGCU---------GAAACCACUUUGUA-------UCCGGAAUCCGGA (((((((((((((((.(((.((.(((((.(((....----)-)).))))).)).))).)))))))))))...)))).---------..............-------(((((...))))) ( -39.40) >DroSec_CAF1 164710 92 + 1 GCCAAGUAUGCAACAGUUUUCGAUUGCUAGCGUCCA----U-GCAGGCGACUGCAAAGAGUUGCAUACUUCUUGGCU---------GAAACCAAUUUGUA-------UCC-------GGA (((((((((((((((((........)))....((..----(-((((....)))))..))))))))))))...)))).---------..............-------...-------... ( -29.80) >DroSim_CAF1 165334 92 + 1 GCCAAGUAUGCAACAGUUUUCGAUUGCCAGCGUCCA----U-GCAGGCGACUGCAAAUAGUUGCAUACUUCUAGGCU---------GAAACCAAUUUGUA-------UCC-------GGA ((((((((((((((.((((.((.(((((.(((....----)-)).))))).)).)))).)))))))))))...))).---------..............-------...-------... ( -31.00) >DroEre_CAF1 168876 105 + 1 GCCAAGUAUGCAACAGUUUUCGAUUGCAAUUG-CCAGCGCUGGCAGGCGACUGCAAAGUGUUGCAUACUUCUGGGCGGAGCGAGCCGAAACGACUUUGGA-------CCC-------GGA ...((((((((((((.(((.((.((((...((-(((....))))).)))).)).))).))))))))))))((((((((((((........)).)))))..-------)))-------)). ( -42.70) >DroYak_CAF1 171257 108 + 1 GCCAAGUAUGCAACAGUUUUCGAUUGCCAGCG-CCA----UGGAUGCUGACUGCCAAGUGUUGCAUACUUCUGGGCUGGGUAAGCCGAAAUGACUUGGGAUACGGAAUCC-------GGA .(((((((((((((.............(((((-(..----..).)))))(((....))))))))))))))...((((.....))))...........((((.....))))-------)). ( -33.70) >consensus GCCAAGUAUGCAACAGUUUUCGAUUGCCAGCGUCCA____U_GCAGGCGACUGCAAAGUGUUGCAUACUUCUGGGCU_________GAAACCACUUUGUA_______UCC_______GGA (((((((((((((((.(((.((.(((((.((...........)).))))).)).))).))))))))))))...)))............................................ (-24.64 = -25.60 + 0.96)

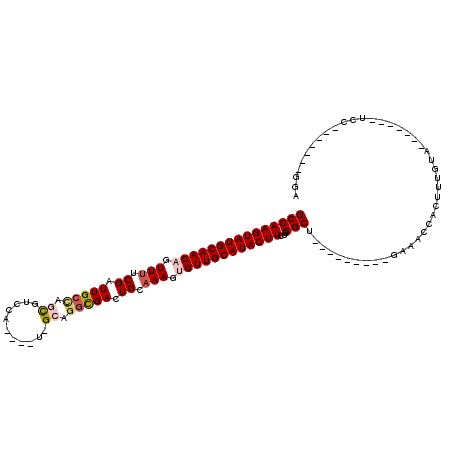
| Location | 8,919,721 – 8,919,820 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Mean single sequence MFE | -32.52 |
| Consensus MFE | -19.72 |
| Energy contribution | -20.60 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8919721 99 - 20766785 UCCGGAUUCCGGA-------UACAAAGUGGUUUC---------AGCCAAGAAGUAUGCAACACUUUGCAGUCGCCUGC-A----UGGACGCUGGCAAUCGAAAACUGUUGCAUACUUGGC (((((...)))))-------.......((((...---------.))))..((((((((((((.(((.(.((.(((.((-.----.....)).))).)).).))).))))))))))))... ( -34.20) >DroSec_CAF1 164710 92 - 1 UCC-------GGA-------UACAAAUUGGUUUC---------AGCCAAGAAGUAUGCAACUCUUUGCAGUCGCCUGC-A----UGGACGCUAGCAAUCGAAAACUGUUGCAUACUUGGC .((-------...-------......(((((...---------.))))).(((((((((((((.((((((.((((...-.----.)).)))).))))..)).....))))))))))))). ( -27.70) >DroSim_CAF1 165334 92 - 1 UCC-------GGA-------UACAAAUUGGUUUC---------AGCCUAGAAGUAUGCAACUAUUUGCAGUCGCCUGC-A----UGGACGCUGGCAAUCGAAAACUGUUGCAUACUUGGC ...-------.((-------.((......)).))---------.(((...(((((((((((..(((.(.((.(((.((-.----.....)).))).)).).)))..)))))))))))))) ( -27.10) >DroEre_CAF1 168876 105 - 1 UCC-------GGG-------UCCAAAGUCGUUUCGGCUCGCUCCGCCCAGAAGUAUGCAACACUUUGCAGUCGCCUGCCAGCGCUGG-CAAUUGCAAUCGAAAACUGUUGCAUACUUGGC .((-------(((-------(....(((((...)))))......))))..((((((((((((..(((((((.(((.((....)).))-).)))))))........)))))))))))))). ( -39.10) >DroYak_CAF1 171257 108 - 1 UCC-------GGAUUCCGUAUCCCAAGUCAUUUCGGCUUACCCAGCCCAGAAGUAUGCAACACUUGGCAGUCAGCAUCCA----UGG-CGCUGGCAAUCGAAAACUGUUGCAUACUUGGC ...-------((((.....)))).(((((.....))))).....(((...((((((((((((.((((..((((((.....----...-.))))))..))))....))))))))))))))) ( -34.50) >consensus UCC_______GGA_______UACAAAGUGGUUUC_________AGCCAAGAAGUAUGCAACACUUUGCAGUCGCCUGC_A____UGGACGCUGGCAAUCGAAAACUGUUGCAUACUUGGC ((((.....))))...............................(((...((((((((((((..(((((((..((..........))..))).))))........))))))))))))))) (-19.72 = -20.60 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:43 2006