
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,919,608 – 8,919,704 |
| Length | 96 |
| Max. P | 0.970332 |

| Location | 8,919,608 – 8,919,704 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -21.98 |
| Consensus MFE | -19.60 |
| Energy contribution | -19.23 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
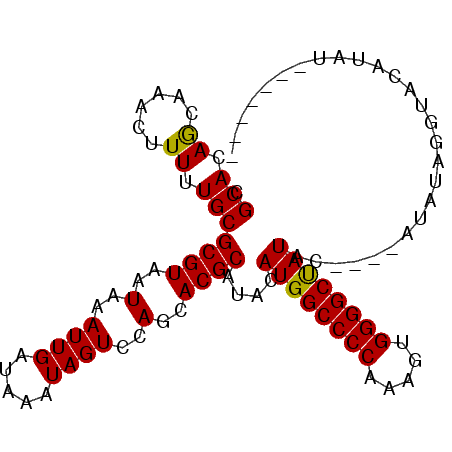
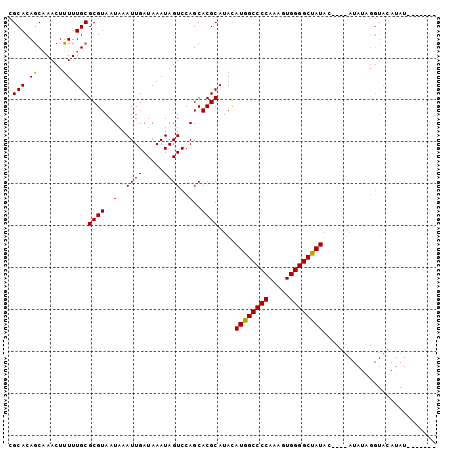
>2R_DroMel_CAF1 8919608 96 + 20766785 CGCACAGCAAACUUUUUGCGCGUAAUAAAUUGAUAAAUAGUCCAGCACGCAUACAUGGCCCCAAAGUGGGGCUAUACUCGUAUAUAGGUACAUAU----A-- ......((((.....))))((((..(..((((.....))))..)..))))....((((((((.....))))))))....((((....))))....----.-- ( -22.50) >DroSec_CAF1 164601 95 + 1 CGCACAGCUAACUUUUUGCGCGUAAUAAAUUGAUAAAUAGUCCAGCACGCAUACAUGGCCCCAAAGUGGGGCUAUAUUCGUAUAUAGGUACAUAU------- ..........((((..(((((((..(..((((.....))))..)..))))....((((((((.....))))))))....)))...))))......------- ( -21.60) >DroSim_CAF1 165231 89 + 1 CGCACAGCAAACUUUUUGCGCGUAAUAAAUUGAUAAAUAGUCCAGCACGCAUACAUGGCCCCAAAGUGGGGCUAU------AUAUUGGUACAUAU------- (((...((((.....))))))).................(((((......(((.((((((((.....))))))))------.)))))).))....------- ( -21.40) >DroEre_CAF1 168752 84 + 1 AGCACAACAAACUUUUUGCGCGUAAUAAAUUGAUAAAUAGUCCAGCACGCAUAUAUGGCCCCAAAGUGGGGCCAUAC------------------AUAUAUC .(((.((......)).)))((((..(..((((.....))))..)..))))...(((((((((.....))))))))).------------------....... ( -22.40) >consensus CGCACAGCAAACUUUUUGCGCGUAAUAAAUUGAUAAAUAGUCCAGCACGCAUACAUGGCCCCAAAGUGGGGCUAUAC____AUAUAGGUACAUAU_______ .(((.((......)).)))((((..(..((((.....))))..)..))))....((((((((.....))))))))........................... (-19.60 = -19.23 + -0.38)

| Location | 8,919,608 – 8,919,704 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 82.17 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -19.49 |
| Energy contribution | -19.30 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
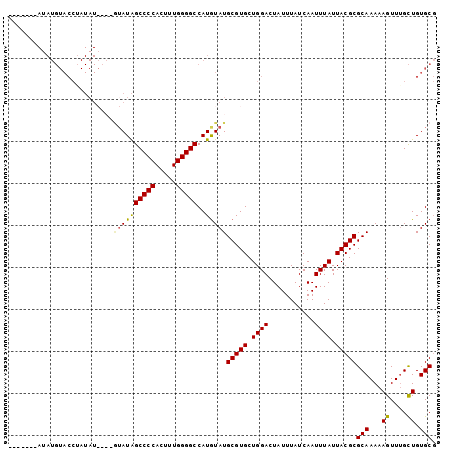
>2R_DroMel_CAF1 8919608 96 - 20766785 --U----AUAUGUACCUAUAUACGAGUAUAGCCCCACUUUGGGGCCAUGUAUGCGUGCUGGACUAUUUAUCAAUUUAUUACGCGCAAAAAGUUUGCUGUGCG --.----....((.((....((((.((((((((((.....)))))..))))).))))..)))).................((((((..........)))))) ( -24.40) >DroSec_CAF1 164601 95 - 1 -------AUAUGUACCUAUAUACGAAUAUAGCCCCACUUUGGGGCCAUGUAUGCGUGCUGGACUAUUUAUCAAUUUAUUACGCGCAAAAAGUUAGCUGUGCG -------....((.((....((((.((((((((((.....)))))..))))).))))..)))).................((((((..........)))))) ( -24.10) >DroSim_CAF1 165231 89 - 1 -------AUAUGUACCAAUAU------AUAGCCCCACUUUGGGGCCAUGUAUGCGUGCUGGACUAUUUAUCAAUUUAUUACGCGCAAAAAGUUUGCUGUGCG -------....((((..((((------((.(((((.....))))).))))))(((((.((((...........)))).)))))((((.....)))).)))). ( -25.90) >DroEre_CAF1 168752 84 - 1 GAUAUAU------------------GUAUGGCCCCACUUUGGGGCCAUAUAUGCGUGCUGGACUAUUUAUCAAUUUAUUACGCGCAAAAAGUUUGUUGUGCU (((((((------------------((((((((((.....))))))))))))).((.....))....))))..........((((((........)))))). ( -29.60) >consensus _______AUAUGUACCUAUAU____GUAUAGCCCCACUUUGGGGCCAUGUAUGCGUGCUGGACUAUUUAUCAAUUUAUUACGCGCAAAAAGUUUGCUGUGCG .........................((((((((((.....)))))..)))))(((((.((((...........)))).)))))(((...((....)).))). (-19.49 = -19.30 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:41 2006