
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,917,513 – 8,917,633 |
| Length | 120 |
| Max. P | 0.724388 |

| Location | 8,917,513 – 8,917,633 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.68 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -30.15 |
| Energy contribution | -30.43 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
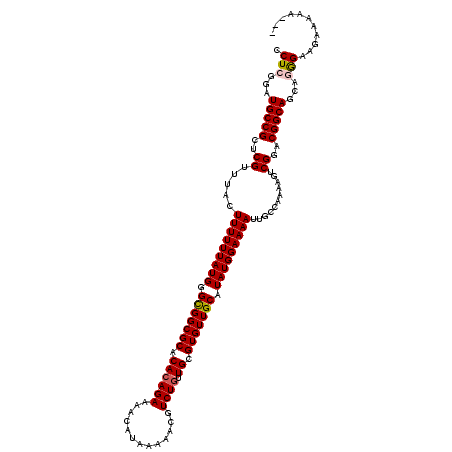
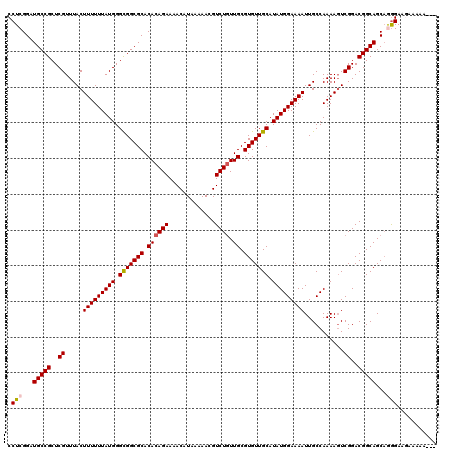
>2R_DroMel_CAF1 8917513 120 + 20766785 CCUCGGAUGCCGCUCGUUUACUUUUUUAUGGGCGGCGCACACAGAAAACAUAAAAACGUCUAUUGCGUGUUGCAUAUGGAAAAUUGCCAAAAGUCGGACGGCAGCAGAGAAGAAAAAAAA .(((....(((((((((..........)))))))))((....................((((((((.....))).)))))....((((...........)))))).)))........... ( -30.20) >DroSec_CAF1 162467 117 + 1 CCUCGGAUGCCGCUCGUUUACUUUUUUAUGGGCGGCGCACACAGAAAACAUAAAAACGUCUGUUGCGUGUUGCAUAUGGAAAAUUGCCAAAACUCGGACGGCAGCUUGGAAGAAAAA--- ((..((.(((((..((.....(((((((((.(((((((.((((((.............)))).)).))))))).)))))))))...........))..))))).)).))........--- ( -31.21) >DroSim_CAF1 163140 117 + 1 CCUCGGAUGCCGCUCGUUUACUUUUUUAUGGGCGGCGCACACAGAAAACAUAAAAACGUCUGUUGCGUGUUGCAUAUGGAAAAUUGCCAAAAGUCGGACGGCAGCAGGGAAGAAAAA--- .(((...(((((..((...(((((.......(((((((.((((((.............)))).)).)))))))...(((.......))))))))))..)))))...)))........--- ( -33.12) >DroEre_CAF1 166702 116 + 1 CCUCGGAUGCCGCUCGUUUACUUUUUUAUGGGUGGCGCACACAGAAAACAUAAAAACGUCUGUUGCGUGUUGCAUAUGGAAAAUUGCCAAAAGUCGGACGGCAGCAAGGAAAAAAA---- (((.(..(((((..((...(((((..((((.(..(((((.(((((.............))))))))))..).))))(((.......))))))))))..))))).).))).......---- ( -31.62) >DroYak_CAF1 168949 116 + 1 CCUCGGAUGCCGCUCGUUUACUUUUUUAUGGGCGGCGCACACAGAAAACAUAAAAACGUCUGUUGCGUGUUGCAUAUGGAAAAUUGCCAAAAGUCGGACGGCAGCAGGGAAAAAAA---- .(((...(((((..((...(((((.......(((((((.((((((.............)))).)).)))))))...(((.......))))))))))..)))))...))).......---- ( -33.12) >consensus CCUCGGAUGCCGCUCGUUUACUUUUUUAUGGGCGGCGCACACAGAAAACAUAAAAACGUCUGUUGCGUGUUGCAUAUGGAAAAUUGCCAAAAGUCGGACGGCAGCAGGGAAGAAAAA___ .(((...(((((..((.....(((((((((.(((((((.((((((.............)))).)).))))))).)))))))))...........))..)))))...)))........... (-30.15 = -30.43 + 0.28)

| Location | 8,917,513 – 8,917,633 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.68 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -28.72 |
| Energy contribution | -28.88 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
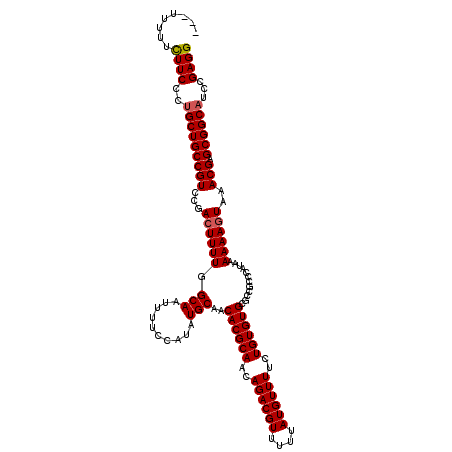
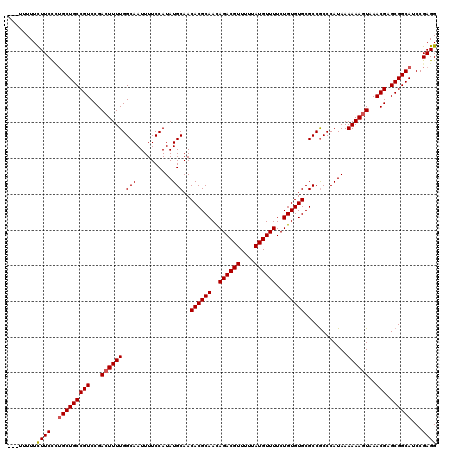
>2R_DroMel_CAF1 8917513 120 - 20766785 UUUUUUUUCUUCUCUGCUGCCGUCCGACUUUUGGCAAUUUUCCAUAUGCAACACGCAAUAGACGUUUUUAUGUUUUCUGUGUGCGCCGCCCAUAAAAAAGUAAACGAGCGGCAUCCGAGG ...........(((.(.(((((..(((((((((((.((.......))((..((((((..((((((....))))))..)))))).)).))).....))))))...))..))))).).))). ( -31.90) >DroSec_CAF1 162467 117 - 1 ---UUUUUCUUCCAAGCUGCCGUCCGAGUUUUGGCAAUUUUCCAUAUGCAACACGCAACAGACGUUUUUAUGUUUUCUGUGUGCGCCGCCCAUAAAAAAGUAAACGAGCGGCAUCCGAGG ---.....((((...(.(((((..((.((..(((.......)))...))..((((((..((((((....))))))..)))))).....................))..))))).).)))) ( -26.80) >DroSim_CAF1 163140 117 - 1 ---UUUUUCUUCCCUGCUGCCGUCCGACUUUUGGCAAUUUUCCAUAUGCAACACGCAACAGACGUUUUUAUGUUUUCUGUGUGCGCCGCCCAUAAAAAAGUAAACGAGCGGCAUCCGAGG ---.....((((..(((((((((...(((((((((.((.......))((..((((((..((((((....))))))..)))))).)).))).....))))))..))).))))))...)))) ( -30.90) >DroEre_CAF1 166702 116 - 1 ----UUUUUUUCCUUGCUGCCGUCCGACUUUUGGCAAUUUUCCAUAUGCAACACGCAACAGACGUUUUUAUGUUUUCUGUGUGCGCCACCCAUAAAAAAGUAAACGAGCGGCAUCCGAGG ----.......(((((.(((((..((((((((.(((..........)))..((((((..((((((....))))))..))))))............))))))...))..)))))..))))) ( -32.00) >DroYak_CAF1 168949 116 - 1 ----UUUUUUUCCCUGCUGCCGUCCGACUUUUGGCAAUUUUCCAUAUGCAACACGCAACAGACGUUUUUAUGUUUUCUGUGUGCGCCGCCCAUAAAAAAGUAAACGAGCGGCAUCCGAGG ----.....(((..(((((((((...(((((((((.((.......))((..((((((..((((((....))))))..)))))).)).))).....))))))..))).))))))...))). ( -29.20) >consensus ___UUUUUCUUCCCUGCUGCCGUCCGACUUUUGGCAAUUUUCCAUAUGCAACACGCAACAGACGUUUUUAUGUUUUCUGUGUGCGCCGCCCAUAAAAAAGUAAACGAGCGGCAUCCGAGG ........((((..(((((((((...((((((.(((..........)))..((((((..((((((....))))))..))))))............))))))..))).))))))...)))) (-28.72 = -28.88 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:40 2006