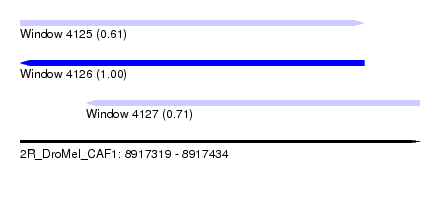
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,917,319 – 8,917,434 |
| Length | 115 |
| Max. P | 0.999914 |

| Location | 8,917,319 – 8,917,418 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -19.84 |
| Consensus MFE | -18.00 |
| Energy contribution | -18.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8917319 99 + 20766785 AUU---------------------GCCCACUUCCCGGACACUGGGCAACUAAAUUAAUUCGAUAAUUGACUUCCAUUGCCUUGCCCGGAACCAUUGUCAUACCUUCCGGAUCUCCAGCUA ...---------------------((.......(((((..((((((((...(((....((((...)))).....)))...)))))))).((....)).......))))).......)).. ( -18.44) >DroSec_CAF1 162281 99 + 1 AUU---------------------GCCCACUUCCCGGACACUGGGCAACUAAAUUAAUUCGAUAAUUGACUUCCAUUGCCUUGCCCGGAUCCAUUGUCAUCCCUUCCGGAUCUCCAGCUA ...---------------------((.......(((((....((((((...(((....((((...)))).....)))...))))))((((........))))..))))).......)).. ( -20.14) >DroSim_CAF1 162954 99 + 1 AUU---------------------GCCCACUUCCCGGACACUGGGCAACUAAAUUAAUUCGAUAAUUGACUUCCAUUGCCUUGCCCGGAACCAUUGUCAUCCCUUCCGGAUCUCCAGCUA ...---------------------((.......(((((..((((((((...(((....((((...)))).....)))...)))))))).((....)).......))))).......)).. ( -18.44) >DroEre_CAF1 166504 99 + 1 AUU---------------------GCCCACUUCCCGGACACUGGGCAACUAAAUUAAUUCGAUAAUUGACUUCCAUUGCCUUGCCCGGAACCAUUGUCAUCCCUUCCGGAUCUCCAGCUC ...---------------------((.......(((((..((((((((...(((....((((...)))).....)))...)))))))).((....)).......))))).......)).. ( -18.44) >DroYak_CAF1 168735 120 + 1 AUUGCCCAUUGCCCAUUGCCCAUUGCCCACUUCCCGGACACUGGGCAACUAAAUUAAUUCGAUAAUUGACUUCCAUUGCCUUGCCCGGAACCAUUGUCAUCCCUUCCGGAUCUCCAGCUC ...((.((..((...(((((((.((.((.......)).)).))))))).....((((((....))))))........))..)).((((((.............)))))).......)).. ( -23.72) >consensus AUU_____________________GCCCACUUCCCGGACACUGGGCAACUAAAUUAAUUCGAUAAUUGACUUCCAUUGCCUUGCCCGGAACCAUUGUCAUCCCUUCCGGAUCUCCAGCUA ........................((.......((((.((..((((((..((.((((((....)))))).))...)))))))).))))...................((....)).)).. (-18.00 = -18.00 + 0.00)

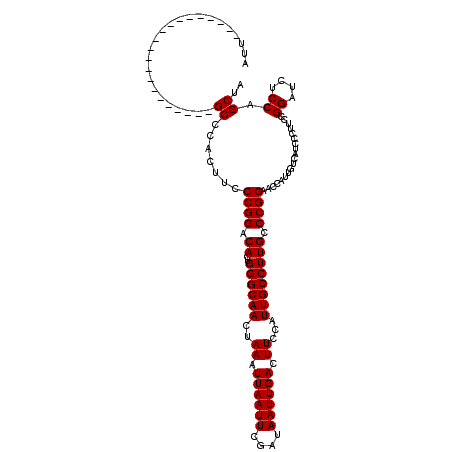

| Location | 8,917,319 – 8,917,418 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.88 |
| Mean single sequence MFE | -33.78 |
| Consensus MFE | -31.20 |
| Energy contribution | -31.60 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.52 |
| SVM RNA-class probability | 0.999914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8917319 99 - 20766785 UAGCUGGAGAUCCGGAAGGUAUGACAAUGGUUCCGGGCAAGGCAAUGGAAGUCAAUUAUCGAAUUAAUUUAGUUGCCCAGUGUCCGGGAAGUGGGC---------------------AAU ..(((......((....)).....((....(((((((((.((((((..((((.((((....)))).)))).))))))...)))))))))..)))))---------------------... ( -32.60) >DroSec_CAF1 162281 99 - 1 UAGCUGGAGAUCCGGAAGGGAUGACAAUGGAUCCGGGCAAGGCAAUGGAAGUCAAUUAUCGAAUUAAUUUAGUUGCCCAGUGUCCGGGAAGUGGGC---------------------AAU ..(((.(...(((....)))...........((((((((.((((((..((((.((((....)))).)))).))))))...))))))))...).)))---------------------... ( -31.90) >DroSim_CAF1 162954 99 - 1 UAGCUGGAGAUCCGGAAGGGAUGACAAUGGUUCCGGGCAAGGCAAUGGAAGUCAAUUAUCGAAUUAAUUUAGUUGCCCAGUGUCCGGGAAGUGGGC---------------------AAU ..(((.....(((....)))....((....(((((((((.((((((..((((.((((....)))).)))).))))))...)))))))))..)))))---------------------... ( -34.00) >DroEre_CAF1 166504 99 - 1 GAGCUGGAGAUCCGGAAGGGAUGACAAUGGUUCCGGGCAAGGCAAUGGAAGUCAAUUAUCGAAUUAAUUUAGUUGCCCAGUGUCCGGGAAGUGGGC---------------------AAU ..(((.....(((....)))....((....(((((((((.((((((..((((.((((....)))).)))).))))))...)))))))))..)))))---------------------... ( -34.00) >DroYak_CAF1 168735 120 - 1 GAGCUGGAGAUCCGGAAGGGAUGACAAUGGUUCCGGGCAAGGCAAUGGAAGUCAAUUAUCGAAUUAAUUUAGUUGCCCAGUGUCCGGGAAGUGGGCAAUGGGCAAUGGGCAAUGGGCAAU ..(((.(.(((((....))).((((..(.(((((......)).))).)..))))....))...........((((((((.((((((.....)))))).))))))))......).)))... ( -36.40) >consensus UAGCUGGAGAUCCGGAAGGGAUGACAAUGGUUCCGGGCAAGGCAAUGGAAGUCAAUUAUCGAAUUAAUUUAGUUGCCCAGUGUCCGGGAAGUGGGC_____________________AAU ..(((.....(((....)))....((....(((((((((.((((((..((((.((((....)))).)))).))))))...)))))))))..)))))........................ (-31.20 = -31.60 + 0.40)


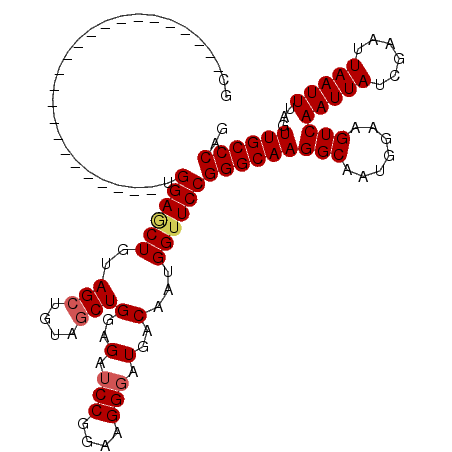
| Location | 8,917,338 – 8,917,434 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.05 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -25.60 |
| Energy contribution | -26.04 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8917338 96 - 20766785 UG------------------------UGGAACUGUAGCUGUAGCUGGAGAUCCGGAAGGUAUGACAAUGGUUCCGGGCAAGGCAAUGGAAGUCAAUUAUCGAAUUAAUUUAGUUGCCCAG ..------------------------.(((((((((((....)))(.....((....)).....).))))))))(((((((((.......)))(((((......)))))...)))))).. ( -27.70) >DroSec_CAF1 162300 96 - 1 GC------------------------UGGAGCUGUAGCUGUAGCUGGAGAUCCGGAAGGGAUGACAAUGGAUCCGGGCAAGGCAAUGGAAGUCAAUUAUCGAAUUAAUUUAGUUGCCCAG .(------------------------((((.((.((((....)))).)).)))))...((((........))))(((((((((.......)))(((((......)))))...)))))).. ( -30.30) >DroSim_CAF1 162973 96 - 1 GC------------------------UGGAACUGUAGCUGUAGCUGGAGAUCCGGAAGGGAUGACAAUGGUUCCGGGCAAGGCAAUGGAAGUCAAUUAUCGAAUUAAUUUAGUUGCCCAG .(------------------------((((.((.((((....)))).)).)))))..(((((.......)))))(((((((((.......)))(((((......)))))...)))))).. ( -30.50) >DroEre_CAF1 166523 114 - 1 GCCGGAGCUGGAGCU------GGAGAUGGAGCUGGAGCUGGAGCUGGAGAUCCGGAAGGGAUGACAAUGGUUCCGGGCAAGGCAAUGGAAGUCAAUUAUCGAAUUAAUUUAGUUGCCCAG (((..((((..((((------........))))..))))(((((((..(.(((....))).)..)...)))))).)))..((((((..((((.((((....)))).)))).))))))... ( -36.70) >DroYak_CAF1 168775 120 - 1 GCUGGUGCUGUUGCUGUUACUGUUGCUGGAGCUGGAGGUGGAGCUGGAGAUCCGGAAGGGAUGACAAUGGUUCCGGGCAAGGCAAUGGAAGUCAAUUAUCGAAUUAAUUUAGUUGCCCAG .((((.(((((((..((..((....(((((.(..(........)..)...))))).))..))..))))))).))))....((((((..((((.((((....)))).)))).))))))... ( -35.40) >consensus GC________________________UGGAGCUGUAGCUGUAGCUGGAGAUCCGGAAGGGAUGACAAUGGUUCCGGGCAAGGCAAUGGAAGUCAAUUAUCGAAUUAAUUUAGUUGCCCAG ...........................((((((..(((....)))(..(.(((....))).)..)...))))))(((((((((.......)))(((((......)))))...)))))).. (-25.60 = -26.04 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:38 2006