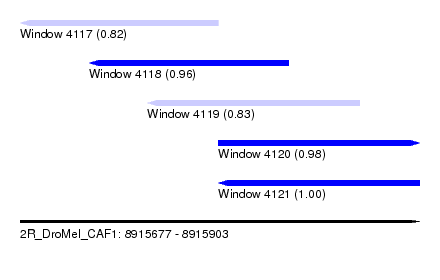
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,915,677 – 8,915,903 |
| Length | 226 |
| Max. P | 0.999634 |

| Location | 8,915,677 – 8,915,789 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.14 |
| Mean single sequence MFE | -38.65 |
| Consensus MFE | -23.99 |
| Energy contribution | -25.18 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
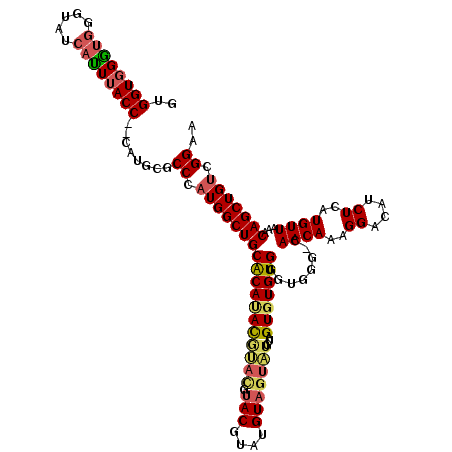
>2R_DroMel_CAF1 8915677 112 - 20766785 GUGGUGGAUGGGUAUCAUUUACCCCCAUAUACCCAUGGCUGCACAUACGUAUGUACAUAUGUA-------GUUUGUGGGUGGG-CAACAAAGGACAUCUCAUGUUAAACAGCUGUCGGAA ...((((..(((((.....)))))))))...((.((((((((((((((((((....)))))))-------...))))((((..-(......)..))))..........))))))).)).. ( -32.30) >DroSec_CAF1 160653 117 - 1 GUGGUGGGUGGGUAUCAUUUACC--CAUGCGCCCAUGGCUGCACAUAUGUACGUACGUAUGUAGUAUGUAGUGUGUGGGUGGG-CAACAAAGGACAUCUCAUGUUAAACAGCUGUCGGAA (.((((.(((((((.....))))--))).)))))((((((((((((((.(((((((.......))))))))))))))((((..-(......)..))))..........)))))))..... ( -43.30) >DroSim_CAF1 161310 117 - 1 GUGGUGGGUGGGUAUCAUUUACC--CAUGCGCCCAUGGCUGCGCAUAUGUACGUACGUAUGUAGUAUGUAGUGUGUGGGUGGG-CAACAAAGGACAUCUCAUGUUAAACAGCUGUCGGAA (.((((.(((((((.....))))--))).)))))((((((((((((((.(((((((.......))))))))))))))((((..-(......)..))))..........)))))))..... ( -42.90) >DroYak_CAF1 167099 105 - 1 -UGGUUGGC-GCUAUCACUUACC--CAUGUGCC--UGGCUGCACACACACAUGU---------GUGUGUAGUGUGUGGGUGCCCCAACAAAGGACAUCUCAUGUUAAACAGCUGUCGGAA -..(((((.-((........(((--((..(...--..(((((((((((....))---------))))))))))..))))))).)))))....((((.((..........)).)))).... ( -36.10) >consensus GUGGUGGGUGGGUAUCAUUUACC__CAUGCGCCCAUGGCUGCACAUACGUACGUACGUAUGUAGUAUGUAGUGUGUGGGUGGG_CAACAAAGGACAUCUCAUGUUAAACAGCUGUCGGAA ..((((((((.....))))))))........((.((((((((((((((((((.(((....)))))))...)))))))........((((..((....))..))))...))))))).)).. (-23.99 = -25.18 + 1.19)

| Location | 8,915,716 – 8,915,829 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.05 |
| Mean single sequence MFE | -42.95 |
| Consensus MFE | -28.73 |
| Energy contribution | -30.72 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
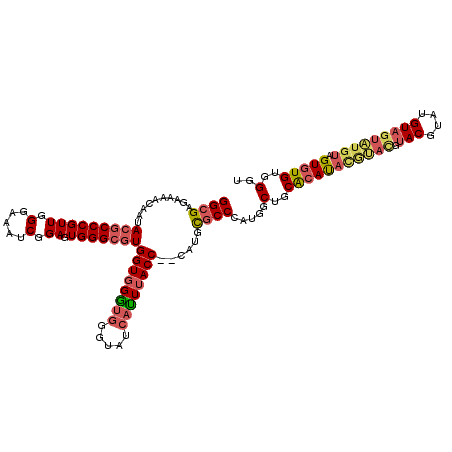
>2R_DroMel_CAF1 8915716 113 - 20766785 GGCGAGAAAACAAUACGCCCGUUGGGAAAUCGGAGUGGGCGUGGUGGAUGGGUAUCAUUUACCCCCAUAUACCCAUGGCUGCACAUACGUAUGUACAUAUGUA-------GUUUGUGGGU .............((((((((((.(.....).)).))))))))((((..(((((.....)))))))))..(((((..((((((..(((....)))....))))-------).)..))))) ( -37.40) >DroSec_CAF1 160692 118 - 1 GGCGAGAAAACAAUACGCCCGUUGGGAAAUCGGAGUGGGCGUGGUGGGUGGGUAUCAUUUACC--CAUGCGCCCAUGGCUGCACAUAUGUACGUACGUAUGUAGUAUGUAGUGUGUGGGU (((...........(((((((((.(.....).)).)))))))((((.(((((((.....))))--))).))))....))).(((((((.(((((((.......))))))))))))))... ( -47.40) >DroSim_CAF1 161349 118 - 1 GGCGAGAAAACAAUACGCCCGUUGGGAAAUCGGAGUGGGCGUGGUGGGUGGGUAUCAUUUACC--CAUGCGCCCAUGGCUGCGCAUAUGUACGUACGUAUGUAGUAUGUAGUGUGUGGGU (((...........(((((((((.(.....).)).)))))))((((.(((((((.....))))--))).))))....))).(((((((.(((((((.......))))))))))))))... ( -47.00) >DroYak_CAF1 167139 104 - 1 GGCGAGAAAACAAUACGCCCGUUGGGAAAUCGGAGUGGG--UGGUUGGC-GCUAUCACUUACC--CAUGUGCC--UGGCUGCACACACACAUGU---------GUGUGUAGUGUGUGGGU ((((......((((.((((((((.(.....).)).))))--)))))).)-))).......(((--((..(...--..(((((((((((....))---------))))))))))..))))) ( -40.00) >consensus GGCGAGAAAACAAUACGCCCGUUGGGAAAUCGGAGUGGGCGUGGUGGGUGGGUAUCAUUUACC__CAUGCGCCCAUGGCUGCACAUACGUACGUACGUAUGUAGUAUGUAGUGUGUGGGU ((((..........(((((((((.(.....).)).)))))))((((((((.....))))))))......)))).....(..(((((((((((.(((....))))))))).)))))..).. (-28.73 = -30.72 + 2.00)


| Location | 8,915,749 – 8,915,869 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.53 |
| Mean single sequence MFE | -40.94 |
| Consensus MFE | -27.42 |
| Energy contribution | -29.94 |
| Covariance contribution | 2.52 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8915749 120 - 20766785 GGCAAAAGUUUUGGUAUAAAACAAAAAUCUGAUUUACAUGGGCGAGAAAACAAUACGCCCGUUGGGAAAUCGGAGUGGGCGUGGUGGAUGGGUAUCAUUUACCCCCAUAUACCCAUGGCU (((.....(((((........))))).((((((((.((((((((...........)))))).))..))))))))(((((..((.(((..(((((.....))))))))))..))))).))) ( -40.30) >DroSec_CAF1 160732 118 - 1 GGCAAAAGUUUUGGUAUAAAACAAAAAUCUGAUUUACAUGGGCGAGAAAACAAUACGCCCGUUGGGAAAUCGGAGUGGGCGUGGUGGGUGGGUAUCAUUUACC--CAUGCGCCCAUGGCU (((.....(((((........))))).((((((((.((((((((...........)))))).))..))))))))(((((((((.(((((((((...)))))))--))))))))))).))) ( -46.70) >DroSim_CAF1 161389 118 - 1 GGCAAAAGUUUUGGUAUAAAACAAAAAUCUGAUUUACAUGGGCGAGAAAACAAUACGCCCGUUGGGAAAUCGGAGUGGGCGUGGUGGGUGGGUAUCAUUUACC--CAUGCGCCCAUGGCU (((.....(((((........))))).((((((((.((((((((...........)))))).))..))))))))(((((((((.(((((((((...)))))))--))))))))))).))) ( -46.70) >DroEre_CAF1 164939 107 - 1 GGCAAAAGUUUUGGUAUAAAACAAAAAUCUGAUUUACAUGGGCGAGAAAACAAUACGCCCGCUGGGAAAUCGGAGUGGG--UGGG-------UAUCAUUUACC--CCUGUGCC--UGGCA .......(((..(((((....((....((((((((.((((((((...........)))))).))..)))))))).))((--.(((-------((.....))))--))))))))--.))). ( -34.10) >DroYak_CAF1 167170 113 - 1 GGCAAAAGUUUUGGAAUAAAACAAAAAUCUGAUUUACAUGGGCGAGAAAACAAUACGCCCGUUGGGAAAUCGGAGUGGG--UGGUUGGC-GCUAUCACUUACC--CAUGUGCC--UGGCU ((((....(((((........))))).((((((((.((((((((...........)))))).))..))))))))(((((--(((.(((.-....))).)))))--))).))))--..... ( -36.90) >consensus GGCAAAAGUUUUGGUAUAAAACAAAAAUCUGAUUUACAUGGGCGAGAAAACAAUACGCCCGUUGGGAAAUCGGAGUGGGCGUGGUGGGUGGGUAUCAUUUACC__CAUGUGCCCAUGGCU .((.....(((((........))))).((((((((.((((((((...........)))))).))..))))))))(((((.((((((((((.....)))))))).....)).))))).)). (-27.42 = -29.94 + 2.52)

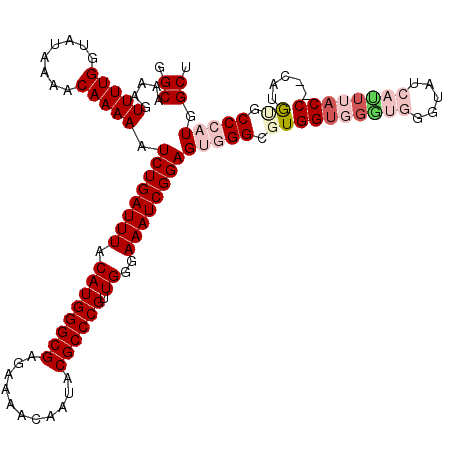

| Location | 8,915,789 – 8,915,903 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.99 |
| Mean single sequence MFE | -37.32 |
| Consensus MFE | -29.62 |
| Energy contribution | -30.22 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8915789 114 + 20766785 GCCCACUCCGAUUUCCCAACGGGCGUAUUGUUUUCUCGCCCAUGUAAAUCAGAUUUUUGUUUUAUACCAAAACUUUUGCCAAUGGAAGUGGAAAGGGAAAU------GGAAUGGGGAUGC .((((.(((.(((((((...(((((...........))))).((((((.(((....))).))))))((...((((((......))))))))...)))))))------))).))))..... ( -36.30) >DroSec_CAF1 160770 114 + 1 GCCCACUCCGAUUUCCCAACGGGCGUAUUGUUUUCUCGCCCAUGUAAAUCAGAUUUUUGUUUUAUACCAAAACUUUUGCCAAUGGAAGUGGAAAGGGAAAU------GGAAUGGGGAUGC .((((.(((.(((((((...(((((...........))))).((((((.(((....))).))))))((...((((((......))))))))...)))))))------))).))))..... ( -36.30) >DroSim_CAF1 161427 114 + 1 GCCCACUCCGAUUUCCCAACGGGCGUAUUGUUUUCUCGCCCAUGUAAAUCAGAUUUUUGUUUUAUACCAAAACUUUUGCCAAUGGAAGUGGAAAGGGAAAU------GGAAUGGGGAUGC .((((.(((.(((((((...(((((...........))))).((((((.(((....))).))))))((...((((((......))))))))...)))))))------))).))))..... ( -36.30) >DroEre_CAF1 164967 119 + 1 -CCCACUCCGAUUUCCCAGCGGGCGUAUUGUUUUCUCGCCCAUGUAAAUCAGAUUUUUGUUUUAUACCAAAACUUUUGCCAGUGGAAGUGGAAGUGGAAAUGGAAAUGGAGUGGGGAUGC -((((((((.((((((....(((((...........))))).((((((.(((....))).))))))((...(((((..(........)..)))))))....))))))))))))))..... ( -41.20) >DroYak_CAF1 167204 119 + 1 -CCCACUCCGAUUUCCCAACGGGCGUAUUGUUUUCUCGCCCAUGUAAAUCAGAUUUUUGUUUUAUUCCAAAACUUUUGCCAAUGGAAGUGGAAAUGGAAAUGAAAUGGGAAUGGGGAUGC -((((.(((.(((((.....(((((...........)))))...........(((((..((((.(((((.............)))))...))))..)))))))))).))).))))..... ( -36.52) >consensus GCCCACUCCGAUUUCCCAACGGGCGUAUUGUUUUCUCGCCCAUGUAAAUCAGAUUUUUGUUUUAUACCAAAACUUUUGCCAAUGGAAGUGGAAAGGGAAAU______GGAAUGGGGAUGC .((((.(((.(((((((...(((((...........))))).((((((.(((....))).))))))((...((((((......))))))))...)))))))......))).))))..... (-29.62 = -30.22 + 0.60)

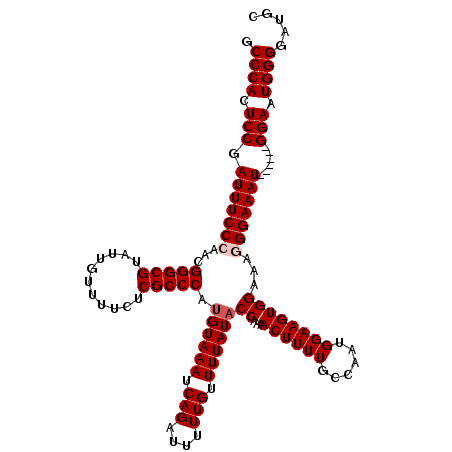
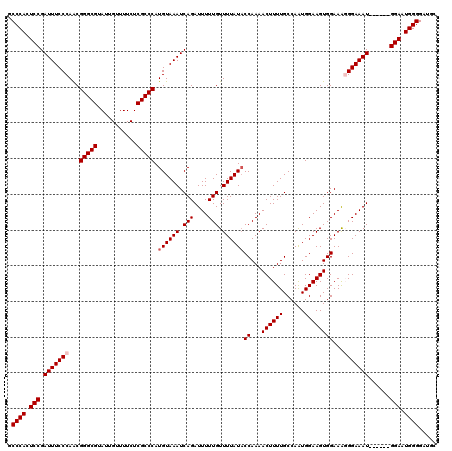
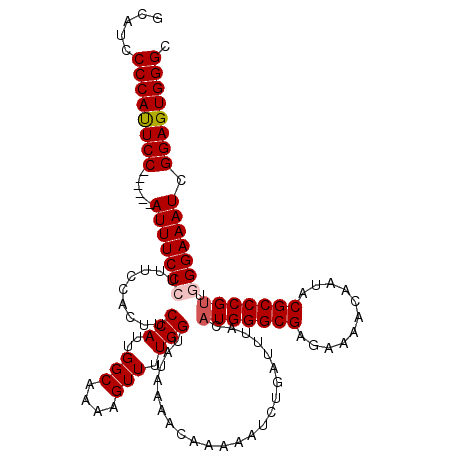
| Location | 8,915,789 – 8,915,903 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.99 |
| Mean single sequence MFE | -35.14 |
| Consensus MFE | -30.10 |
| Energy contribution | -30.54 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.81 |
| SVM RNA-class probability | 0.999634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8915789 114 - 20766785 GCAUCCCCAUUCC------AUUUCCCUUUCCACUUCCAUUGGCAAAAGUUUUGGUAUAAAACAAAAAUCUGAUUUACAUGGGCGAGAAAACAAUACGCCCGUUGGGAAAUCGGAGUGGGC .....((((((((------(((((((.........(((..(((....))).))).......................(((((((...........))))))).))))))).)))))))). ( -35.80) >DroSec_CAF1 160770 114 - 1 GCAUCCCCAUUCC------AUUUCCCUUUCCACUUCCAUUGGCAAAAGUUUUGGUAUAAAACAAAAAUCUGAUUUACAUGGGCGAGAAAACAAUACGCCCGUUGGGAAAUCGGAGUGGGC .....((((((((------(((((((.........(((..(((....))).))).......................(((((((...........))))))).))))))).)))))))). ( -35.80) >DroSim_CAF1 161427 114 - 1 GCAUCCCCAUUCC------AUUUCCCUUUCCACUUCCAUUGGCAAAAGUUUUGGUAUAAAACAAAAAUCUGAUUUACAUGGGCGAGAAAACAAUACGCCCGUUGGGAAAUCGGAGUGGGC .....((((((((------(((((((.........(((..(((....))).))).......................(((((((...........))))))).))))))).)))))))). ( -35.80) >DroEre_CAF1 164967 119 - 1 GCAUCCCCACUCCAUUUCCAUUUCCACUUCCACUUCCACUGGCAAAAGUUUUGGUAUAAAACAAAAAUCUGAUUUACAUGGGCGAGAAAACAAUACGCCCGCUGGGAAAUCGGAGUGGG- .....((((((((((((((............(((...(((......)))...))).....................((((((((...........)))))).)))))))).))))))))- ( -35.20) >DroYak_CAF1 167204 119 - 1 GCAUCCCCAUUCCCAUUUCAUUUCCAUUUCCACUUCCAUUGGCAAAAGUUUUGGAAUAAAACAAAAAUCUGAUUUACAUGGGCGAGAAAACAAUACGCCCGUUGGGAAAUCGGAGUGGG- .....((((((((.(((((....(((.......(((((..(((....))).))))).....................(((((((...........))))))))))))))).))))))))- ( -33.10) >consensus GCAUCCCCAUUCC______AUUUCCCUUUCCACUUCCAUUGGCAAAAGUUUUGGUAUAAAACAAAAAUCUGAUUUACAUGGGCGAGAAAACAAUACGCCCGUUGGGAAAUCGGAGUGGGC .....((((((((......(((((((.........(((..(((....))).))).......................(((((((...........))))))).))))))).)))))))). (-30.10 = -30.54 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:32 2006