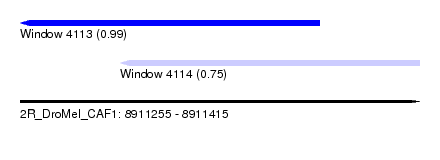
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,911,255 – 8,911,415 |
| Length | 160 |
| Max. P | 0.990728 |

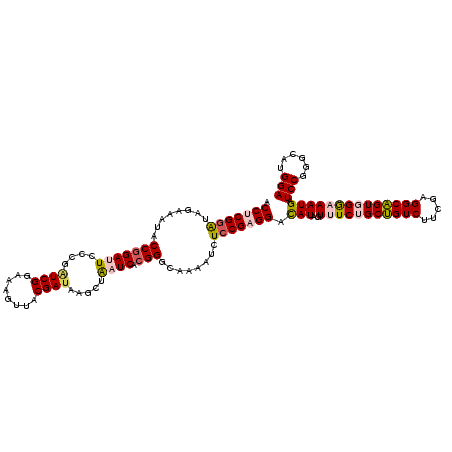
| Location | 8,911,255 – 8,911,375 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.33 |
| Mean single sequence MFE | -43.68 |
| Consensus MFE | -39.24 |
| Energy contribution | -39.20 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8911255 120 - 20766785 GAUAAGCUAAUCACGGGCAAAAUCUCCGAGGACAUAGUUUCUGCUGUCUUCGAGGCAGUGGGCAAUGAUCCGGGCAUGGACCGCUUACUGGCGGUUCUGGAGAGGAAUCGCAAGUGUGAA ..........(((((.((....((((((.((((((.((..(..(((((.....)))))..))).))).)))......((((((((....))))))))))))))......))...))))). ( -42.70) >DroSec_CAF1 156249 120 - 1 GAUAAGCUAAUCACGGGCAAAAUCUCCGAGGACAUAGUUUCUGCUGUCUUCGAGGCAGUGGAAAAUGAUCCCGGCAUGGACCGCUUACUGGCGGUUCUGGAGAGGAAUCGCAAGUGCGCA .....((....(((..((....((((((.((((((..((((..(((((.....)))))..))))))).)))).....((((((((....)))))))).)))))......))..))).)). ( -45.10) >DroSim_CAF1 156821 120 - 1 GAUAAGCUAAUCACGGGCAAAAUCUCCGAGGACAUAGUUUCUGCUGUCUUCGAGGCAGUGGAAAAUGAUCCCGGCAUGGACCGCUUACUGGCGGUUCUGGAGAGGAAUCGCAAGUGCGCA .....((....(((..((....((((((.((((((..((((..(((((.....)))))..))))))).)))).....((((((((....)))))))).)))))......))..))).)). ( -45.10) >DroEre_CAF1 160561 120 - 1 GAUAAGCUGAUCACGGACAAGAUCUCCGAGGACAUAGUUUCCGCCGUCUUCGAGGCGGUUGGCAAUGAUCCGGGCAUGGACCGCUUGCUGGCGGUUCUGGAGAGGAAUCGCAAGUGCGCG .....(((.....((((.......)))).((((((.(((.(((((........)))))..))).))).))).)))......(((.((((.((((((((.....)))))))).)))).))) ( -43.00) >DroYak_CAF1 162593 120 - 1 GAUAAACUAAUCACGGGCAAAAUUUCCGAGGAUAUAGUUUCCGCUGUCUUCGAGGCAGUUGGAAAUGAUCCGAGCAUGGAUCGCUUGCUGGCGGUUCUGGAGAGGAAUCGCAAGUGUGCG (((.......((.((((.......)))).((((...((((((((((((.....))))).))))))).))))))......)))((.((((.((((((((.....)))))))).)))).)). ( -42.52) >consensus GAUAAGCUAAUCACGGGCAAAAUCUCCGAGGACAUAGUUUCUGCUGUCUUCGAGGCAGUGGGAAAUGAUCCGGGCAUGGACCGCUUACUGGCGGUUCUGGAGAGGAAUCGCAAGUGCGCA ...........(((..((....((((((.((((((..(((((((((((.....)))))))))))))).)))......((((((((....))))))))))))))......))..))).... (-39.24 = -39.20 + -0.04)

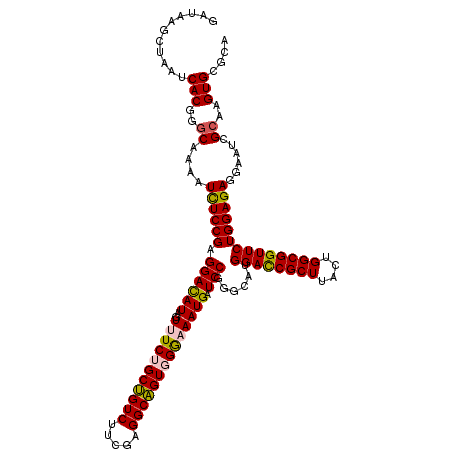
| Location | 8,911,295 – 8,911,415 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.33 |
| Mean single sequence MFE | -38.41 |
| Consensus MFE | -30.03 |
| Energy contribution | -31.07 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8911295 120 - 20766785 ACCUCGGAUUGAAAUACCGGAUUCCCGAUCGGAAAGUUACGAUAAGCUAAUCACGGGCAAAAUCUCCGAGGACAUAGUUUCUGCUGUCUUCGAGGCAGUGGGCAAUGAUCCGGGCAUGGA .(((((((........(((((((....((((........)))).....)))).)))........))))))).(((.((..(..(((((.....)))))..))).))).((((....)))) ( -37.79) >DroSec_CAF1 156289 120 - 1 ACCUCGGAUAGAAAUACCGGAUUCCCGAUCGGAAAGUUACGAUAAGCUAAUCACGGGCAAAAUCUCCGAGGACAUAGUUUCUGCUGUCUUCGAGGCAGUGGAAAAUGAUCCCGGCAUGGA .(((((((........(((((((....((((........)))).....)))).)))........))))))).(((..((((..(((((.....)))))..))))))).(((......))) ( -38.79) >DroSim_CAF1 156861 120 - 1 ACCUCGGAUAGAAAUACCGGAUUCCCGAUCGGAAAGUUACGAUAAGCUAAUCACGGGCAAAAUCUCCGAGGACAUAGUUUCUGCUGUCUUCGAGGCAGUGGAAAAUGAUCCCGGCAUGGA .(((((((........(((((((....((((........)))).....)))).)))........))))))).(((..((((..(((((.....)))))..))))))).(((......))) ( -38.79) >DroEre_CAF1 160601 120 - 1 ACCACGGGUGGAAAUACCAGAGUCCCGCUCGGAAAGCUACGAUAAGCUGAUCACGGACAAGAUCUCCGAGGACAUAGUUUCCGCCGUCUUCGAGGCGGUUGGCAAUGAUCCGGGCAUGGA .(((....))).....(((..((((...(((((.((((......))))((((........)))))))))((((((.(((.(((((........)))))..))).))).))))))).))). ( -42.90) >DroYak_CAF1 162633 120 - 1 ACCACGAAUAGAAAUACCGGAGUCGCAGUCGGAAAGUUACGAUAAACUAAUCACGGGCAAAAUUUCCGAGGAUAUAGUUUCCGCUGUCUUCGAGGCAGUUGGAAAUGAUCCGAGCAUGGA .(((((..((....)).)).....((..((((((((((..(((......)))...)))....)))))))((((...((((((((((((.....))))).))))))).))))..)).))). ( -33.80) >consensus ACCUCGGAUAGAAAUACCGGAUUCCCGAUCGGAAAGUUACGAUAAGCUAAUCACGGGCAAAAUCUCCGAGGACAUAGUUUCUGCUGUCUUCGAGGCAGUGGGAAAUGAUCCGGGCAUGGA .(((((((........(((((((....((((........)))).....)))).)))........))))))).(((..(((((((((((.....)))))))))))))).(((......))) (-30.03 = -31.07 + 1.04)

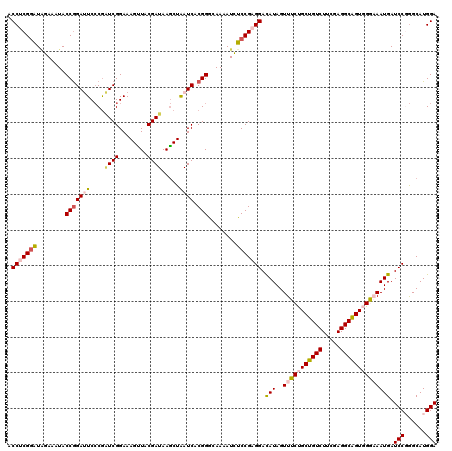
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:25 2006