| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,910,935 – 8,911,095 |
| Length | 160 |
| Max. P | 0.827373 |

| Location | 8,910,935 – 8,911,055 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -39.60 |
| Consensus MFE | -29.58 |
| Energy contribution | -30.50 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8910935 120 + 20766785 AACUUCUUGACAAUUCUCGACUUUUGGCUGAUCUUACUCGGAUCGAGCAAUCGCACAUCCUGACUGAAAUCCUCGGCAAGGAUUUGAAGUGUGCUCUGUCGCUUCUCACGACACUUUUGU .........((((...(((......(((.(((((.....)))))(((((....((.(((((..((((.....))))..))))).)).....)))))....))).....))).....)))) ( -28.80) >DroSec_CAF1 155929 120 + 1 AACUUCUUGACGAUCCUCGACUUUUGGCUGAUCUUAGUCGGAUCGAGCAAUCGCACAUCCUGGCUGAAAUCCUCGGCAAGGAUUUGAAGUGUACUCUGUCGCUUCUCGCGACACUUUUGC .(((((.((.((((.(((((..((((((((....)))))))))))))..)))))).(((((.(((((.....))))).)))))..)))))......((((((.....))))))....... ( -40.30) >DroSim_CAF1 156501 120 + 1 AACUUCUUGACGAUCCUCGACUUUUGGCUGAUCUUAGUCGGAUCGAGCAAUCGCACAUCCUGGCUGAAAUCCUCGGCAAGGAUUUGAAGUGUGCUCUGUCGCUUCUCGCGACACUUUUGC ...........(((((..((((...((....))..)))))))))(((((....((.(((((.(((((.....))))).))))).)).....)))))((((((.....))))))....... ( -40.40) >DroEre_CAF1 160241 120 + 1 AACUUCUUGACGAUCCUCGACUUUUGGUUGAUCUUACUUGGAUCGAGUAAACGCACAUCCUGGCUGAAGUCCUCGGCCAGGAUUUGAAGUGAGCUCUGUCGCUUCUCGCGGCACUUGCGC .(((((....(((((((((((.....)))))........))))))...........(((((((((((.....)))))))))))..)))))..((..((((((.....))))))...)).. ( -45.30) >DroYak_CAF1 162273 120 + 1 AACUUCUUGACGAUCCUGGACUUUUGGCUGAUCUUGCUCGGAUCGAGCAAUCGCACAUCCUGGCUGAAAUCCUCGGCCAGGAGUUGAAGUGAGCUCUGUCGCUUCUCGCGACACUUUCGC .(((((..(((.............((((.(((..((((((...))))))))))).))((((((((((.....))))))))))))))))))(((...((((((.....))))))..))).. ( -43.20) >consensus AACUUCUUGACGAUCCUCGACUUUUGGCUGAUCUUACUCGGAUCGAGCAAUCGCACAUCCUGGCUGAAAUCCUCGGCAAGGAUUUGAAGUGUGCUCUGUCGCUUCUCGCGACACUUUUGC .............................(((((.....)))))(((((....((.(((((.(((((.....))))).))))).)).....)))))((((((.....))))))....... (-29.58 = -30.50 + 0.92)

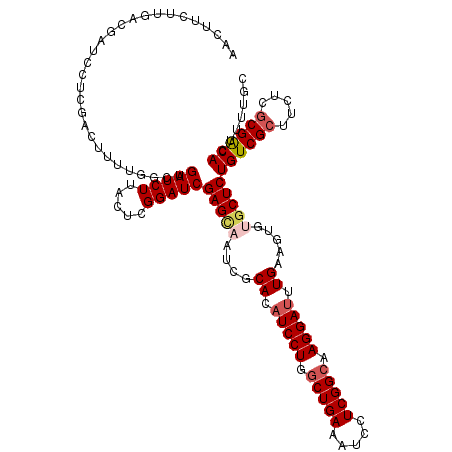
| Location | 8,910,975 – 8,911,095 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.75 |
| Mean single sequence MFE | -40.51 |
| Consensus MFE | -30.30 |
| Energy contribution | -30.10 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8910975 120 + 20766785 GAUCGAGCAAUCGCACAUCCUGACUGAAAUCCUCGGCAAGGAUUUGAAGUGUGCUCUGUCGCUUCUCACGACACUUUUGUUGCUCGUUGGAUGUCUUGAGAAUUAGGUCAUAGAAGUCCU (((.(((((....((.(((((..((((.....))))..))))).)).....))))).)))..((((((.((((((..((.....))..)).)))).))))))..(((.(......).))) ( -36.90) >DroSec_CAF1 155969 120 + 1 GAUCGAGCAAUCGCACAUCCUGGCUGAAAUCCUCGGCAAGGAUUUGAAGUGUACUCUGUCGCUUCUCGCGACACUUUUGCUGCUCGUUCGAUGUCUUGAGAAUUAGGUCAUAGAAGUCCU ((.((((((...(((((((((.(((((.....))))).))))).....))))....((((((.....)))))).......)))))).))(((.((((((........))).))).))).. ( -38.40) >DroSim_CAF1 156541 120 + 1 GAUCGAGCAAUCGCACAUCCUGGCUGAAAUCCUCGGCAAGGAUUUGAAGUGUGCUCUGUCGCUUCUCGCGACACUUUUGCUGCUCGUUAGAUGUCUUGAGAAUUAGGUCAUAGAAGUCCU (((.(((((....((.(((((.(((((.....))))).))))).)).....))))).)))..((((((.((((((...((.....)).)).)))).))))))..(((.(......).))) ( -38.40) >DroEre_CAF1 160281 120 + 1 GAUCGAGUAAACGCACAUCCUGGCUGAAGUCCUCGGCCAGGAUUUGAAGUGAGCUCUGUCGCUUCUCGCGGCACUUGCGCUCCUCGUUGGACGUCUUCAGUAUCAGGUCAUAGAAGUCCU ...((((......((.(((((((((((.....))))))))))).))....((((.(((((((.....))))))...).))))))))..((((.(((...............))).)))). ( -43.16) >DroYak_CAF1 162313 120 + 1 GAUCGAGCAAUCGCACAUCCUGGCUGAAAUCCUCGGCCAGGAGUUGAAGUGAGCUCUGUCGCUUCUCGCGACACUUUCGCUCCUCGUUGGACGUCUUUAGGAUUAGGUCGUAGAAGUCCU (((.((((..((((...((((((((((.....))))))))))......)))))))).)))((((((.(((((.((.....((((....((....))..))))..)))))))))))))... ( -45.70) >consensus GAUCGAGCAAUCGCACAUCCUGGCUGAAAUCCUCGGCAAGGAUUUGAAGUGUGCUCUGUCGCUUCUCGCGACACUUUUGCUGCUCGUUGGAUGUCUUGAGAAUUAGGUCAUAGAAGUCCU (((.(((((....((.(((((.(((((.....))))).))))).)).....))))).)))((((((...(((...(((......(((...)))......)))....)))..))))))... (-30.30 = -30.10 + -0.20)

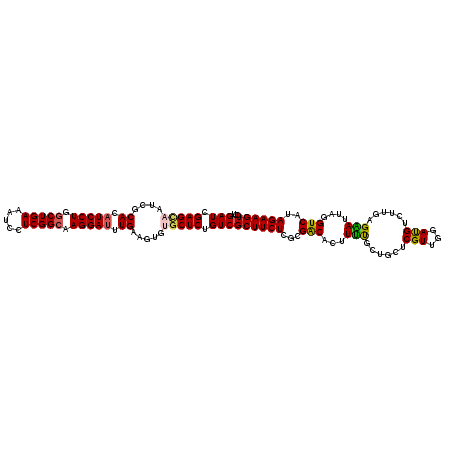
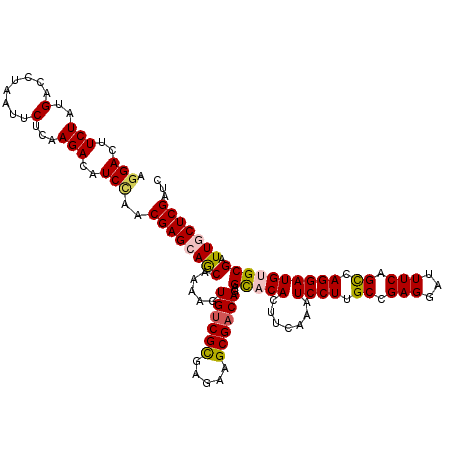
| Location | 8,910,975 – 8,911,095 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.75 |
| Mean single sequence MFE | -38.28 |
| Consensus MFE | -28.53 |
| Energy contribution | -28.97 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.654101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8910975 120 - 20766785 AGGACUUCUAUGACCUAAUUCUCAAGACAUCCAACGAGCAACAAAAGUGUCGUGAGAAGCGACAGAGCACACUUCAAAUCCUUGCCGAGGAUUUCAGUCAGGAUGUGCGAUUGCUCGAUC .(((..(((.(((........))))))..)))..((((((((.....((((((.....))))))..((((((((.((((((((...)))))))).....))).)))))).)))))))... ( -37.30) >DroSec_CAF1 155969 120 - 1 AGGACUUCUAUGACCUAAUUCUCAAGACAUCGAACGAGCAGCAAAAGUGUCGCGAGAAGCGACAGAGUACACUUCAAAUCCUUGCCGAGGAUUUCAGCCAGGAUGUGCGAUUGCUCGAUC ..((..(((.(((........))))))..))...((((((((.....((((((.....))))))..((((((((.((((((((...)))))))).....))).)))))).)))))))... ( -34.10) >DroSim_CAF1 156541 120 - 1 AGGACUUCUAUGACCUAAUUCUCAAGACAUCUAACGAGCAGCAAAAGUGUCGCGAGAAGCGACAGAGCACACUUCAAAUCCUUGCCGAGGAUUUCAGCCAGGAUGUGCGAUUGCUCGAUC .(((..(((.(((........))))))..)))..((((((((.....((((((.....))))))..((((((((.((((((((...)))))))).....))).)))))).)))))))... ( -37.00) >DroEre_CAF1 160281 120 - 1 AGGACUUCUAUGACCUGAUACUGAAGACGUCCAACGAGGAGCGCAAGUGCCGCGAGAAGCGACAGAGCUCACUUCAAAUCCUGGCCGAGGACUUCAGCCAGGAUGUGCGUUUACUCGAUC .((((.(((...............))).))))..(((((((((((((((.(....).(((......)))))))....((((((((.(((...))).)))))))).))))))).))))... ( -40.66) >DroYak_CAF1 162313 120 - 1 AGGACUUCUACGACCUAAUCCUAAAGACGUCCAACGAGGAGCGAAAGUGUCGCGAGAAGCGACAGAGCUCACUUCAACUCCUGGCCGAGGAUUUCAGCCAGGAUGUGCGAUUGCUCGAUC .((((.(((...............))).))))..((((((((.....((((((.....))))))..))))...((.(((((((((.(((...))).))))))).))..))...))))... ( -42.36) >consensus AGGACUUCUAUGACCUAAUUCUCAAGACAUCCAACGAGCAGCAAAAGUGUCGCGAGAAGCGACAGAGCACACUUCAAAUCCUUGCCGAGGAUUUCAGCCAGGAUGUGCGAUUGCUCGAUC .(((..(((..(........)...)))..)))..((((((((.....((((((.....))))))..(((((.......((((.((.(((...))).)).)))))))))).)))))))... (-28.53 = -28.97 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:22 2006