
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,897,311 – 8,897,410 |
| Length | 99 |
| Max. P | 0.566520 |

| Location | 8,897,311 – 8,897,410 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 70.09 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -5.81 |
| Energy contribution | -7.98 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.19 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8897311 99 + 20766785 GCAGUUUUAGCCAGCUG---GCGUUUGUUA--UAUUACGG--AAGUCUGUUAGCCAACCUG-GCUAAAUCAGAGUGAGAGGCACGUUCAUGCCUCAAUGCAUCAUAA ((((.(((((((((.((---((........--....((((--....))))..))))..)))-)))))).).......((((((......))))))..)))....... ( -33.34) >DroVir_CAF1 186789 95 + 1 ACAGUUG---CCG-UUG---CAUAUAACGAACUUUGCGGCGCAAGU-UGUGUGGCAGCAUGUGUUGAAUCAAAGUCA----UGCAUUUAUGGUUCAAUGCAAUGUGU (((((((---(((-(((---....)))).......(((((....))-)))..)))))).((((((((((((((((..----...)))).)))))))))))).))).. ( -31.00) >DroSec_CAF1 142149 99 + 1 GCAGUUCUAGCCAGCUG---GAGUUAGCUA--UAUUCUGG--AAGUCUGUUAGCCAUCCUG-GCUAAAUCAGAGUCUGAGGCACGUUCAUGCCUCAAUGCAUCAUAA (((.((((((..(((((---....))))).--....))))--)).((((((((((.....)-)))))..))))...(((((((......))))))).)))....... ( -31.90) >DroSim_CAF1 142270 99 + 1 GCAGUUCUAGCCAGCUG---GAGUUAGCUA--UAUUCUGG--AAGUGUGUUAGCCAUCCUG-GCUAAAUCAGAGUCUGAGGCACGUUCAUGCCUCAAUGCAUCAUAA (((....(((((((...---(.((((((.(--((((....--.))))))))))))...)))-))))..........(((((((......))))))).)))....... ( -31.80) >DroYak_CAF1 148381 99 + 1 ACACUUCUAGCCAGCUG---CAGUUUGCUU--UAUGCUGG--AAGAAUGUUAGCCAUCCUG-GCUAAAUCAGAGUCUGAGGCACGUUCAUGCCUCAAUGCAUCAUAA ...((((((((.(((..---......))).--...)))))--)))....((((((.....)-)))))....((((.(((((((......)))))))..)).)).... ( -32.70) >DroAna_CAF1 142000 91 + 1 UGAGUUCUGGCUGGGGGCGAGUCUUGGCCA--C---UUGA--AUGUGUGGUAACCAACCUG-GCUAAAUCAAAGU--------CGCUCAUGGCUCAAUGCAUUGUAA ...((..(((((..((((((.(.(((((((--(---....--....)))))..((.....)-)......)))).)--------)))))..))).))..))....... ( -25.00) >consensus GCAGUUCUAGCCAGCUG___GAGUUAGCUA__UAUUCUGG__AAGUCUGUUAGCCAUCCUG_GCUAAAUCAGAGUCUGAGGCACGUUCAUGCCUCAAUGCAUCAUAA ........(((.((((.....)))).)))....................(((((........)))))......((..((((((......))))))...))....... ( -5.81 = -7.98 + 2.17)

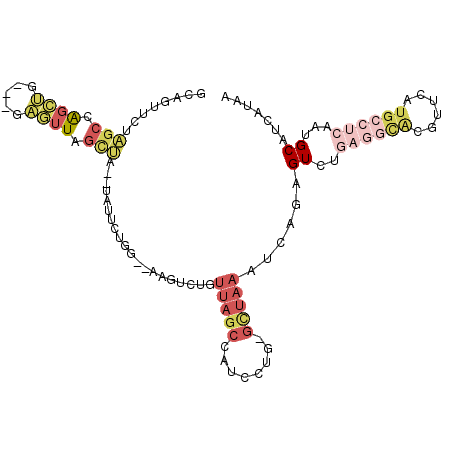
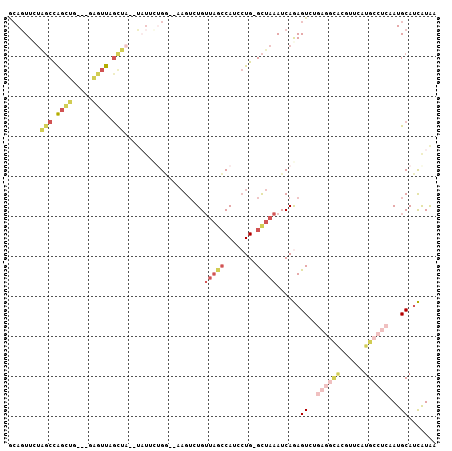
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:12 2006