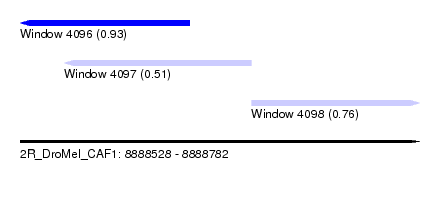
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,888,528 – 8,888,782 |
| Length | 254 |
| Max. P | 0.929077 |

| Location | 8,888,528 – 8,888,636 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -19.48 |
| Energy contribution | -21.32 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8888528 108 - 20766785 GUUGCUAAAAAUUGGCAGCUUAAAAUGAAGUAUAUAUUUUAUAUAACUUUAGGAUAUGUGCCACAAACAGCGGCUGUUGAUUCCAACAAAGGGCAAACAAAGGCCAAU ((((.....(((..((((((.....((..((((((((((((........))))))))))))..))......))))))..))).))))....(((........)))... ( -28.40) >DroSec_CAF1 133304 108 - 1 GUUGCUAAAAAUUGGCAGCUCAACAUGAGGUAUAUAUAUUAUAUAUCUUUAGGUUAUGUACCACAAAAAGCGGCUGUUGAAUCCAAUUAAGGGCAAACAAAGGCCAUU (((((((.....)))))))((((((.((((((((((...))))))))))..(((.....)))............))))))...........(((........)))... ( -26.10) >DroSim_CAF1 133156 108 - 1 GUUGCUAAAAAUUGGCAGCUCAACAUGAGGUAUAACUUUUAUAUAUCUUUAGGAUAUGUGCCACAAAAAGCGGCUGUUGAUUCCAAUUAAGGGCAAACAAAGGCCAUU (((((((.....)))))))((((((...............(((((((.....)))))))(((.(.....).)))))))))...........(((........)))... ( -26.40) >DroEre_CAF1 136595 107 - 1 GUUGCUAAAAAUUGCCAACUUGACAUA-GGUAUAUAUUUGAUAUAUCUUUGGGAUAUGCGCCACAAAAAGCGGCCGUUGAUGCCAAUUAAGGGCAAACAAAGGCCGUA (((((........).))))........-((..(((((((.(........).)))))))..)).......((((((.(((.((((.......))))..))).)))))). ( -29.50) >DroYak_CAF1 138665 108 - 1 GUAGCUAAAAAUUGCCAGCUUAACAUAAGGUAUAUAUUUUAUAUAUCUUUAGGAUAUGUGCCACAAAAAGUAGCCGUUGAUUCUAAUCAAGAGCAAACUAAAGCGUUU ...((........))..((((.......(((((((((((((........)))))))))))))..........((..(((((....)))))..))......)))).... ( -24.10) >consensus GUUGCUAAAAAUUGGCAGCUUAACAUGAGGUAUAUAUUUUAUAUAUCUUUAGGAUAUGUGCCACAAAAAGCGGCUGUUGAUUCCAAUUAAGGGCAAACAAAGGCCAUU (((((((.....))))))).........(((((((((((((........))))))))))))).........((((.(((...((.......))....))).))))... (-19.48 = -21.32 + 1.84)


| Location | 8,888,556 – 8,888,675 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.77 |
| Mean single sequence MFE | -26.72 |
| Consensus MFE | -21.44 |
| Energy contribution | -22.92 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.80 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8888556 119 - 20766785 GCUAAUAUUCAUGAGUAUUUAUAUAGGGCGCGC-UUAAAAGUUGCUAAAAAUUGGCAGCUUAAAAUGAAGUAUAUAUUUUAUAUAACUUUAGGAUAUGUGCCACAAACAGCGGCUGUUGA ...((((((....)))))).......(((.(((-(...(((((((((.....)))))))))....((..((((((((((((........))))))))))))..))...)))))))..... ( -29.40) >DroSec_CAF1 133332 119 - 1 ACCAAUAUUCAUGAGUAUUUAUUUAGGGCGCGC-UAAAAUGUUGCUAAAAAUUGGCAGCUCAACAUGAGGUAUAUAUAUUAUAUAUCUUUAGGUUAUGUACCACAAAAAGCGGCUGUUGA ..((((.....(((((....))))).(((.(((-(.....(((((((.....)))))))......((.((((((((..(((........)))..)))))))).))...))))))))))). ( -27.50) >DroSim_CAF1 133184 119 - 1 ACCAAUAUUUAUGAGUAUUUAUUUAGGGCGCGC-UAAAAUGUUGCUAAAAAUUGGCAGCUCAACAUGAGGUAUAACUUUUAUAUAUCUUUAGGAUAUGUGCCACAAAAAGCGGCUGUUGA ..((((.....(((((....))))).(((.(((-(.....(((((((.....)))))))......((.((((((..(((((........)))))..)))))).))...))))))))))). ( -26.00) >DroEre_CAF1 136623 118 - 1 AGCAAUAUUCAUGAGCACUUAUUUAGGGCGCGC-UAAAAUGUUGCUAAAAAUUGCCAACUUGACAUA-GGUAUAUAUUUGAUAUAUCUUUGGGAUAUGCGCCACAAAAAGCGGCCGUUGA (((((((((...(.((.(((.....))).)).)-...))))))))).........((((....((.(-(((((((.....)))))))).))((...(((..........))).)))))). ( -27.50) >DroYak_CAF1 138693 120 - 1 ACCAAUAUUCAUGAGCAUUUAUUUAGGGCGCGAACAAAAUGUAGCUAAAAAUUGCCAGCUUAACAUAAGGUAUAUAUUUUAUAUAUCUUUAGGAUAUGUGCCACAAAAAGUAGCCGUUGA ..((((.....(((((....((((..(((....((.....)).)))..)))).....)))))......(((((((((((((........))))))))))))).............)))). ( -23.20) >consensus ACCAAUAUUCAUGAGUAUUUAUUUAGGGCGCGC_UAAAAUGUUGCUAAAAAUUGGCAGCUUAACAUGAGGUAUAUAUUUUAUAUAUCUUUAGGAUAUGUGCCACAAAAAGCGGCUGUUGA .........................((.(((.........(((((((.....))))))).........(((((((((((((........))))))))))))).......))).))..... (-21.44 = -22.92 + 1.48)

| Location | 8,888,675 – 8,888,782 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.12 |
| Mean single sequence MFE | -24.64 |
| Consensus MFE | -18.15 |
| Energy contribution | -17.55 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8888675 107 + 20766785 GGAGAGCUAUAAAUCCCCUUUGAAAAGGCAUAUAU----AUAUAUUCACCUGUGCCCCAUCCA------CAUCCACUCCAUUAACGUGGGUUUUAUGGCCCACUGGCGCCACACCA-U ((...((((.................((((((...----...........)))))).......------................(((((((....))))))))))).))......-. ( -22.84) >DroSec_CAF1 133451 107 + 1 GGAUAGCUAUAAAUCCCCUUUGGAAAGGCAUAUAU----AUAUAUUCACCUGUGCCCCACCCA------CAUCCACUCCAUUAACGUGGAUUUUAUGGCCUACUGGCGCCACACCA-U ((...((((((((.......(((...((((((...----...........))))))....)))------.((((((.........))))))))))))))....((....))..)).-. ( -22.44) >DroSim_CAF1 133303 111 + 1 GGAUAGCUAUAAAUCCCCUUUGGAAAGGCAUAUAUAUAUAUAUAUUCACCUGUGCCCCACCCA------CAUCCACUCCAUUAACGUGGAUUUUAUGGCCUACUGGCGCCACACCA-U ((((........))))....(((...(((((((((....))))))...((.(((..(((....------.((((((.........))))))....)))..))).)).)))...)))-. ( -23.60) >DroEre_CAF1 136741 104 + 1 GUAUAGCUGUAAAUCCCCUUUGGAGAGGCAUAU--------ACAUUCACCUGUGCCACAUCCA------CACCCACUCCAUUAACGUGGGUUUUAUGGCCUACUGGCGCCCCACCACU ((((((((.....(((.....)))..))).)))--------))........((((((...(((------.((((((.........))))))....))).....))))))......... ( -27.70) >DroYak_CAF1 138813 109 + 1 GGAUAGCUAUAAAUCCCCUUUGGAAAGGCAUAU--------AUAUUUAGCUGUGCCACACCCACAUCCACAGCUACUCCAUUAACCUGGGUUUUAUGGCCUACUGGCGCCCCACCA-U ((((........))))....(((...(((....--------.....((((((((.............))))))))...(((.(((....)))..)))(((....))))))...)))-. ( -26.62) >consensus GGAUAGCUAUAAAUCCCCUUUGGAAAGGCAUAUAU____AUAUAUUCACCUGUGCCCCACCCA______CAUCCACUCCAUUAACGUGGGUUUUAUGGCCUACUGGCGCCACACCA_U .....((((((((........((...((((((..................))))))...)).........((((((.........))))))))))))))....(((.......))).. (-18.15 = -17.55 + -0.60)


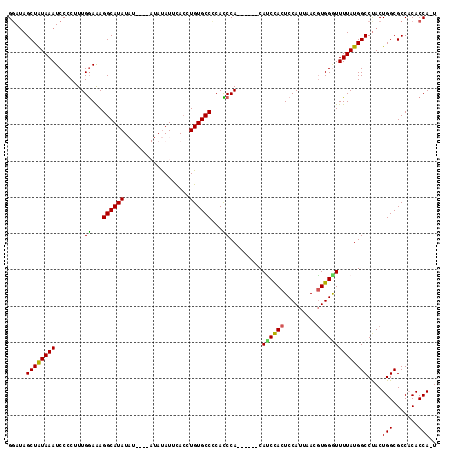
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:31:09 2006