
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,873,533 – 8,873,780 |
| Length | 247 |
| Max. P | 0.999851 |

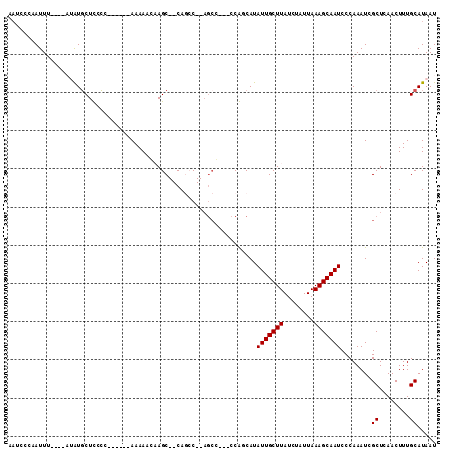
| Location | 8,873,533 – 8,873,628 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.15 |
| Mean single sequence MFE | -16.04 |
| Consensus MFE | -7.00 |
| Energy contribution | -7.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.44 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519641 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8873533 95 - 20766785 AAUCCCAAUUU----AUAUGCUCACC------AAAAACAAGC--CAGCC--AGCG---CCGGCAUAUUGCUUAUCUAUUAAAGCAAUCCCAAAUCGCUCAACUUUGCAUAAU ...........----.(((((.....------........((--(.((.--...)---).)))..(((((((........)))))))..................))))).. ( -17.20) >DroPse_CAF1 131444 97 - 1 UAUCCCAAUUU------GUGCUUCCC------AAAAUGCAGCUCCAGACGAAGCC---CCACCAUAUUGCUUAUCUAUUAAAGCAAUCCCAAAUCGCUCAACUUUGCAUAAU .........((------((((.....------.....((.(((.(....).))).---.......(((((((........)))))))........))........)))))). ( -13.67) >DroSim_CAF1 118174 95 - 1 AAUCCCAAUUU----AUAUGCUCACC------AAAAACAAGC--CAGCC--AGCG---CCGGCAUAUUGCUUAUCUAUUAAAGCAAUCCCAAAUCGCUCAACUUUGCAUAAU ...........----.(((((.....------........((--(.((.--...)---).)))..(((((((........)))))))..................))))).. ( -17.20) >DroYak_CAF1 122846 99 - 1 AAUCCCCAUUUACAUAUAUGCUCCCC------AAAAACAAGC--CAACC--AGGC---CCGGCAUAUUGCUUAUCUAUUAAAGCAAUCCCAAAUCGCUCAACUUUGCAUAAU ................(((((.....------........((--(....--.)))---..((...(((((((........))))))).))...............))))).. ( -16.00) >DroAna_CAF1 117697 102 - 1 --CACCAAUUU----AUAUCCUUGGCUUGUACAAAAACAAGC--CAUCC--UUCGACAUCAACAUAUUGCUUAUCUAUUAAAGCAAUCCCAAAUCGCUCAACUUUGCAUAAU --.........----.......((((((((......))))))--))...--..............(((((((........)))))))........((........))..... ( -18.50) >DroPer_CAF1 134300 97 - 1 UAUCCCAAUUU------GUGCUUCCC------AAAAUGCAGCUCCAGACGAAGCC---CCACCAUAUUGCUUAUCUAUUAAAGCAAUCCCAAAUCGCUCAACUUUGCAUAAU .........((------((((.....------.....((.(((.(....).))).---.......(((((((........)))))))........))........)))))). ( -13.67) >consensus AAUCCCAAUUU____AUAUGCUCCCC______AAAAACAAGC__CAGCC__AGCC___CCAGCAUAUUGCUUAUCUAUUAAAGCAAUCCCAAAUCGCUCAACUUUGCAUAAU .................................................................(((((((........)))))))........((........))..... ( -7.00 = -7.00 + 0.00)


| Location | 8,873,601 – 8,873,708 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.88 |
| Mean single sequence MFE | -22.02 |
| Consensus MFE | -12.05 |
| Energy contribution | -11.92 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
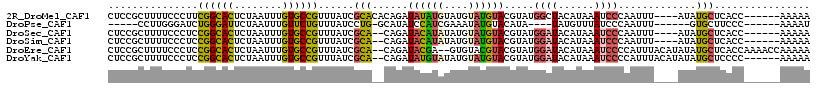
>2R_DroMel_CAF1 8873601 107 - 20766785 CUCCGCUUUUCCCUUCGGCACUCUAAUUUGUGCCGUUUAUCGCACACAGAUAUAUGUAUGUAUGUACGUAUGGCUACAUAAAUCCCAAUUU----AUAUGCUCACC------AAAAA ...............((((((........))))))......(((...((.(((((((((....))))))))).))..((((((....))))----)).))).....------..... ( -21.30) >DroPse_CAF1 131516 95 - 1 -----CCUUGGGAUCUGGGAUUCUAAUUUGUUCUGUUUAUCCUG-GCAUAUCCAUCGAAAUAUGUACAUA----UAUGUUUAUCCCAAUUU------GUGCUUCCC------AAAAU -----..((((((...(((((...(((.......))).)))))(-(((((.......((((((((....)----))))))).........)------)))))))))------))... ( -20.69) >DroSec_CAF1 118396 105 - 1 CUCCGCUUUUCCCUCCGGCACUCUAAUUUGUGCCGUUUAUCGCA--CAGAUACAUAUAUGUAUGUACGUAUGGAUACAUAAAUCCCAAUUU----AUAUGCUCACC------AAAAA ...............((((((........))))))..((((...--..)))).......(((((((.((..((((......))))..)).)----)))))).....------..... ( -20.70) >DroSim_CAF1 118242 105 - 1 CUCCGCUUUUCCCUCCGGCACUCUAAUUUGUGCCGUUUAUCGCA--CAGAUACAUAUAUGUAUGUACGUAUGGAUACAUAAAUCCCAAUUU----AUAUGCUCACC------AAAAA ...............((((((........))))))..((((...--..)))).......(((((((.((..((((......))))..)).)----)))))).....------..... ( -20.70) >DroEre_CAF1 119946 113 - 1 CUCCGCUUUUCCCUCCGGCACUCUAAUUUGUGCCGUUUAUCGCA--CAGAUACGA--GUGUACGUACGUAUGGAUACAUAAAUCCCCAUUUACAUAUAUGCUCACCAAAACCAAAAA ....((...........((((((..((((((((........)))--)))))..))--))))..(((.((..((((......))))..)).)))......))................ ( -23.80) >DroYak_CAF1 122914 109 - 1 CUCCGCUUUUCCCUCCGGCACUCUAAUUUGUGCCGUUUAUCGCA--CAGAUAUGUAUAUGUAUGUACGUAUGGAUACAUAAAUCCCCAUUUACAUAUAUGCUCCCC------AAAAA ....((.........((((((........))))))..((((...--..)))).....(((((((((.((..((((......))))..)).))))))))))).....------..... ( -24.90) >consensus CUCCGCUUUUCCCUCCGGCACUCUAAUUUGUGCCGUUUAUCGCA__CAGAUACAUAUAUGUAUGUACGUAUGGAUACAUAAAUCCCAAUUU____AUAUGCUCACC______AAAAA ...............((((((........))))))......(((......((((((....)))))).....((((......)))).............)))................ (-12.05 = -11.92 + -0.13)
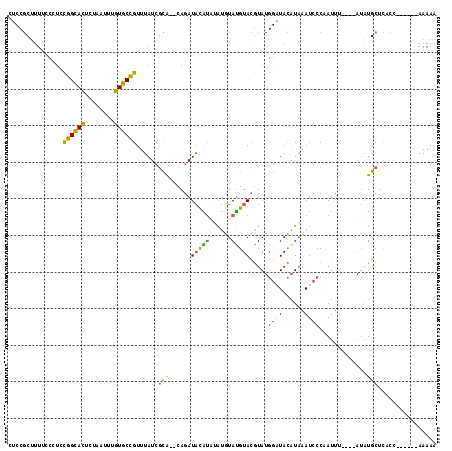
| Location | 8,873,668 – 8,873,780 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 95.10 |
| Mean single sequence MFE | -31.96 |
| Consensus MFE | -29.68 |
| Energy contribution | -29.72 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.25 |
| SVM RNA-class probability | 0.999851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
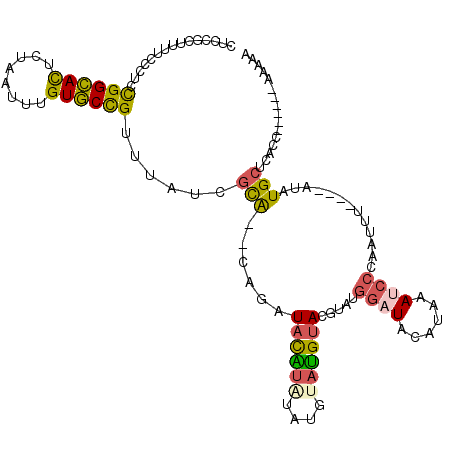
>2R_DroMel_CAF1 8873668 112 + 20766785 AUAAACGGCACAAAUUAGAGUGCCGAAGGGAAAAGCGGAGACAGCAAAUCAUCGGUGGCAGAUUGCUCUAAGGGAAACCGGAAAAUUGCUCGAAAAUGCCUUCAAG------AUACAG ......(((((........)))))(((((.(...(((....).))......(((..(((((.((..(((..((....))))).))))))))))...).)))))...------...... ( -31.30) >DroSec_CAF1 118461 112 + 1 AUAAACGGCACAAAUUAGAGUGCCGGAGGGAAAAGCGGAGACAGCAAAUCAUCGGUGGCAGAUUGCUCUAAGGGAAACCGGAAAAUUGCUCGAAAAUGCCUUCAAG------AUACAG .....((((((........))))))(((((....(((....).))......(((..(((((.((..(((..((....))))).)))))))))).....)))))...------...... ( -32.60) >DroSim_CAF1 118307 112 + 1 AUAAACGGCACAAAUUAGAGUGCCGGAGGGAAAAGCGGAGACAGCAAAUCAUCGGUGGCAGAUUGCUCUAAGGAAAACCGGAAAAUUGCUCGAAAAUGCCUUCAAG------AUACAG .....((((((........))))))(((((......(....)(((((....((((((((.....))).........)))))....)))))........)))))...------...... ( -29.70) >DroEre_CAF1 120019 112 + 1 AUAAACGGCACAAAUUAGAGUGCCGGAGGGAAAAGCGGAGACGGCAAAUCAUCGGUGGCAGAUUGCUCUGAGGGAAACCGGAAAAUUGCUCGAAAAUGCCUUUAAG------AUACAG .....((((((........))))))(((((.....((....))((((....((((.(((.....)))))))((....))......)))).........)))))...------...... ( -33.90) >DroYak_CAF1 122983 118 + 1 AUAAACGGCACAAAUUAGAGUGCCGGAGGGAAAAGCGGAGACAGCAAAUCAUCGGUGGCAGAUUGCUCAGAGGGAAACCGGAAAAUUGCUCGAAAAUGCCUUUAAUGCCUUUAUACAG .....((((((........))))))(((((....(((....).))....(((..(.((((....((.....((....))........)).......)))).)..))))))))...... ( -32.32) >consensus AUAAACGGCACAAAUUAGAGUGCCGGAGGGAAAAGCGGAGACAGCAAAUCAUCGGUGGCAGAUUGCUCUAAGGGAAACCGGAAAAUUGCUCGAAAAUGCCUUCAAG______AUACAG .....((((((........))))))(((((....(((....).))......(((..(((((.((..((...((....)).)).)))))))))).....)))))............... (-29.68 = -29.72 + 0.04)

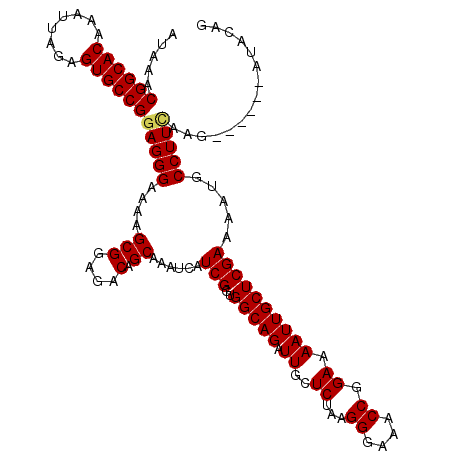
| Location | 8,873,668 – 8,873,780 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 95.10 |
| Mean single sequence MFE | -27.94 |
| Consensus MFE | -27.46 |
| Energy contribution | -27.02 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.02 |
| SVM RNA-class probability | 0.999758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8873668 112 - 20766785 CUGUAU------CUUGAAGGCAUUUUCGAGCAAUUUUCCGGUUUCCCUUAGAGCAAUCUGCCACCGAUGAUUUGCUGUCUCCGCUUUUCCCUUCGGCACUCUAAUUUGUGCCGUUUAU ......------...((((((.....(.(((((.((..((((......((((....))))..))))..)).)))))).....)))))).....((((((........))))))..... ( -27.40) >DroSec_CAF1 118461 112 - 1 CUGUAU------CUUGAAGGCAUUUUCGAGCAAUUUUCCGGUUUCCCUUAGAGCAAUCUGCCACCGAUGAUUUGCUGUCUCCGCUUUUCCCUCCGGCACUCUAAUUUGUGCCGUUUAU ......------...((((((.....(.(((((.((..((((......((((....))))..))))..)).)))))).....)))))).....((((((........))))))..... ( -27.60) >DroSim_CAF1 118307 112 - 1 CUGUAU------CUUGAAGGCAUUUUCGAGCAAUUUUCCGGUUUUCCUUAGAGCAAUCUGCCACCGAUGAUUUGCUGUCUCCGCUUUUCCCUCCGGCACUCUAAUUUGUGCCGUUUAU ......------...((((((.....(.(((((.((..((((......((((....))))..))))..)).)))))).....)))))).....((((((........))))))..... ( -27.60) >DroEre_CAF1 120019 112 - 1 CUGUAU------CUUAAAGGCAUUUUCGAGCAAUUUUCCGGUUUCCCUCAGAGCAAUCUGCCACCGAUGAUUUGCCGUCUCCGCUUUUCCCUCCGGCACUCUAAUUUGUGCCGUUUAU ......------...((((((.....((.((((.((..((((......((((....))))..))))..)).)))))).....)))))).....((((((........))))))..... ( -30.50) >DroYak_CAF1 122983 118 - 1 CUGUAUAAAGGCAUUAAAGGCAUUUUCGAGCAAUUUUCCGGUUUCCCUCUGAGCAAUCUGCCACCGAUGAUUUGCUGUCUCCGCUUUUCCCUCCGGCACUCUAAUUUGUGCCGUUUAU ........(((....((((((.....(.(((((.((..((((........((....))....))))..)).)))))).....)))))).))).((((((........))))))..... ( -26.60) >consensus CUGUAU______CUUGAAGGCAUUUUCGAGCAAUUUUCCGGUUUCCCUUAGAGCAAUCUGCCACCGAUGAUUUGCUGUCUCCGCUUUUCCCUCCGGCACUCUAAUUUGUGCCGUUUAU ...............((((((.....((.((((.((..((((......((((....))))..))))..)).)))))).....)))))).....((((((........))))))..... (-27.46 = -27.02 + -0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:57 2006