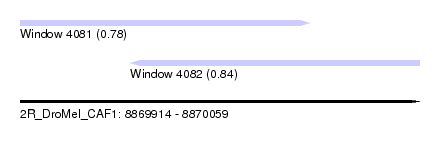
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,869,914 – 8,870,059 |
| Length | 145 |
| Max. P | 0.841530 |

| Location | 8,869,914 – 8,870,019 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.77 |
| Mean single sequence MFE | -37.44 |
| Consensus MFE | -28.38 |
| Energy contribution | -28.18 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8869914 105 + 20766785 AUAGUGCUACGAAAAGUUACCCAUGGAGCACUUACAUCGUGGAACUGCCUGGACACCUGGCCAUCCAUGUCCAGGCUGCUCCGACAGAUUGGGCAACGCUUGGCC .....((((.(....(((.(((((((((((((........))....(((((((((..(((....)))))))))))))))))))......)))).))).).)))). ( -38.10) >DroSec_CAF1 114764 94 + 1 CU-----------AAGUUGCCCAUGGAGCACUUACAUCGUGGAACUGCCUGGACACCCGGUCACCCAUGUCCAGGCUGCACCGACAGAUUGGGCAGCGCUUUGCC ..-----------..(((((((((((.(((((........))....(((((((((...((....)).)))))))))))).)))......))))))))........ ( -35.40) >DroSim_CAF1 114588 94 + 1 CU-----------AAGUUGCCCAUGGAGCACUUACAUCGUGGAACUGCCUGGACACCCGGUCACCCAUGUCCAGGCUGCACCGACAGAUUGGGCAGCGCUUUGCC ..-----------..(((((((((((.(((((........))....(((((((((...((....)).)))))))))))).)))......))))))))........ ( -35.40) >DroEre_CAF1 116259 96 + 1 CUAG---------AAGCUGCUCAUGGAGCACUUACAUAGUGCAACUGCCUGGACACCCGCUCCUCCAUGUCCAGGCGACGACGGCAGAUGGGGCAGCGCUUGGCC ((((---------..(((((((.((..(((((.....)))))...((((((((((............)))))))))).......))....)))))))..)))).. ( -38.00) >DroYak_CAF1 119155 103 + 1 CUAGUGUU--UAAACGUUGCUCAUGGAGCACUUACAUCGCGCAACCGCCUGGGCACCUGGUCGUCCAUGUCCAGGCGGCGCCGGCAGAUGGGACAACGCUUGGCC (.((((((--.......(((((...)))))....((((((((..(((((((((((..(((....)))))))))))))).))..)).))))....)))))).)... ( -40.30) >consensus CUAG_________AAGUUGCCCAUGGAGCACUUACAUCGUGGAACUGCCUGGACACCCGGUCAUCCAUGUCCAGGCUGCACCGACAGAUUGGGCAGCGCUUGGCC ...............((((((((((.........))).((((....(((((((((...((....)).)))))))))....)).)).....)))))))........ (-28.38 = -28.18 + -0.20)

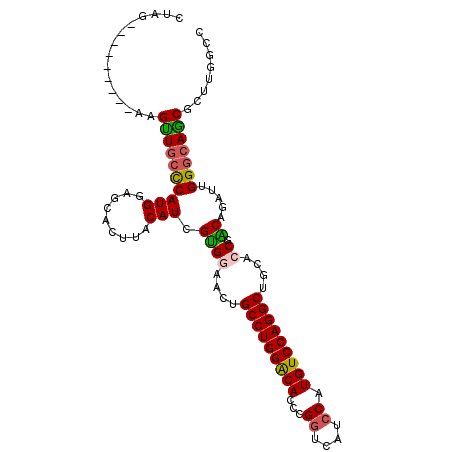
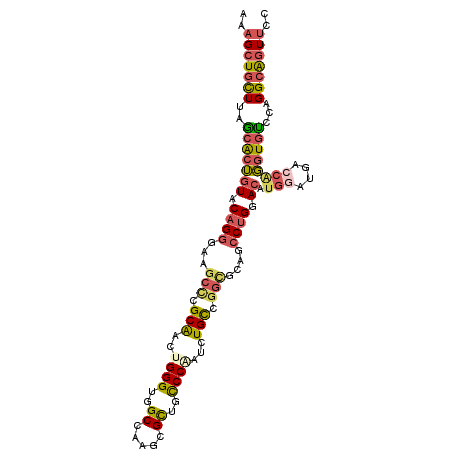
| Location | 8,869,954 – 8,870,059 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 80.32 |
| Mean single sequence MFE | -46.78 |
| Consensus MFE | -30.64 |
| Energy contribution | -31.03 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8869954 105 - 20766785 AAAGCUGUUUAGCACUGUACAGGGAAGCUCGCAACUGGGUGGCCAAGCGUUGCCCAAUCUGUCGGAGCAGCCUGGACAUGGAUGGCCAGGUGUCCAGGCAGUUCC ....((((.((....)).))))(((((((((((..((((..((.....))..))))...)))..)))).((((((((((((....)))..)))))))))..)))) ( -43.20) >DroSec_CAF1 114793 105 - 1 AAAGCUGCUUAGCACUGUACAGGGAAGCCCGCAGCUGGGUGGCAAAGCGCUGCCCAAUCUGUCGGUGCAGCCUGGACAUGGGUGACCGGGUGUCCAGGCAGUUCC ..((((.....((((((.(((((..(((.....)))(((..((.....))..)))..))))))))))).((((((((((((....))..)))))))))))))).. ( -54.00) >DroSim_CAF1 114617 105 - 1 AAAGCUGCUUAGCACUGUACAGGGAAGCCCGCAGCUGGGUGGCAAAGCGCUGCCCAAUCUGUCGGUGCAGCCUGGACAUGGGUGACCGGGUGUCCAGGCAGUUCC ..((((.....((((((.(((((..(((.....)))(((..((.....))..)))..))))))))))).((((((((((((....))..)))))))))))))).. ( -54.00) >DroEre_CAF1 116290 105 - 1 AAAGCUGCUUGGCACUGUACAGGGAAGCCCGCAACUGGGUGGCCAAGCGCUGCCCCAUCUGCCGUCGUCGCCUGGACAUGGAGGAGCGGGUGUCCAGGCAGUUGC ..(((((((((((.((....))....((((((....(((..((.....))..)))..(((.((((.(((.....))))))).)))))))))).)))))))))).. ( -47.70) >DroYak_CAF1 119193 105 - 1 AAAGCUGCUUAGCACUGUACAGGGAAGCCCGCAACUGGGUGGCCAAGCGUUGUCCCAUCUGCCGGCGCCGCCUGGACAUGGACGACCAGGUGCCCAGGCGGUUGC ..((((((((.(((((((.(((((..(((.(((..((((((((.....)))).))))..))).)))..).)))).)).(((....))))))))..)))))))).. ( -41.90) >DroPer_CAF1 130310 99 - 1 AGGGCUGCUUGCAGAAAUACCGCGAGGCGCGCGGAUGGGUGGCCGAGCGCUGUCCGCUCUGCCGGCGCAGACUGGA------UGAUUAUGUGGCCCGGAACUGGA .((((..(((((.........))))(((((.(((........))).)))))(((((.(((((....))))).))))------)......)..))))......... ( -39.90) >consensus AAAGCUGCUUAGCACUGUACAGGGAAGCCCGCAACUGGGUGGCCAAGCGCUGCCCAAUCUGCCGGCGCAGCCUGGACAUGGAUGACCAGGUGUCCAGGCAGUUCC ..(((((((..(((((((.((((...(((.(((..((((..((.....))..))))...))).)))....)))).)).(((....))))))))...))))))).. (-30.64 = -31.03 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:54 2006