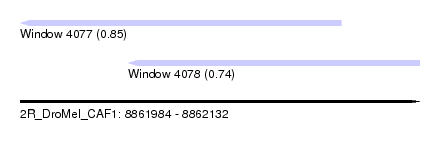
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,861,984 – 8,862,132 |
| Length | 148 |
| Max. P | 0.851679 |

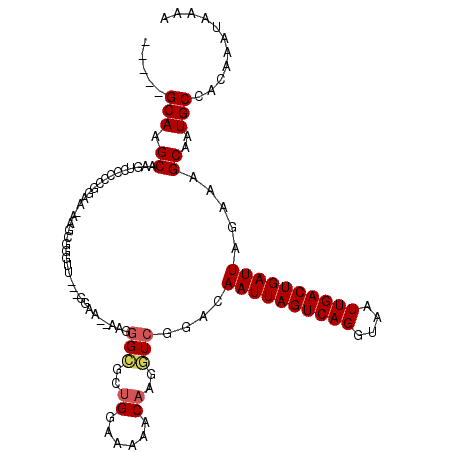
| Location | 8,861,984 – 8,862,103 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.58 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -12.46 |
| Energy contribution | -12.70 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8861984 119 - 20766785 GGAAA-AAGGGCGCUGGAAAAACAAGGUCGGACAAUUAGUCAGGUAAUUGACUGAUUAGAAAGCAAUGCCAGAAAUAAAAUGGCUAAUGUUUGAUGUAAUGUUUAAUUUUCAUUUAACAG (((((-..(((((.((......))(.(((((((((((((((((....))))))))))..........((((.........))))....))))))).)..)))))..)))))......... ( -25.00) >DroSec_CAF1 106866 99 - 1 GGAA--AAGGGCGCUGGAAAAACAAGGUCGGACAAUUAGUCAGGUAAUUGACUGAUUAGAAAGCAAUGCCACAAAUAAAA-------------------UGUUUGAUUUUCAUUUAAGCG ....--.....((((..(((...((((((((((((((((((((....)))))))))).....((...))...........-------------------.))))))))))..))).)))) ( -20.80) >DroSim_CAF1 106777 99 - 1 GGAA--AAGGGCGCUGGAAAAACAAGGUCGGACAAUUAGUCAGGUAAUUGACUGAUUAGAAAGCAAUGCCACAAAUGAAA-------------------UGUUUAAUUUCCAUUUAAUCG ((((--(..((((.((.........(......)((((((((((....))))))))))......)).))))..((((....-------------------.))))..)))))......... ( -19.50) >DroEre_CAF1 108533 105 - 1 ----------GGGUUGGAAAAACCAGCUCGG-CAAUUAGUCAGGUAAUUGACUGAUUGCAAAGCAAUGCCACAAAUUAAAGUGCCAAUGUUUGCUGUAAUAGUUAAUUCUCACUUC---- ----------(((((((.....))))))).(-(((((((((((....)))))))))))).(((((.((.(((........))).)).)))))........................---- ( -32.30) >DroYak_CAF1 110969 119 - 1 GGAAAAAGGGGCGCUGGAAAAACAAGCUCGG-CAAUUAGUCAGGUAAUUGACUGAUUAGAAAGCAAUGCCACAAAUAAAAGUGCCAAUGUUUACAGCCAUGGUUAAUUGUCACUCUAGUG ...........(((((((...(((((((.((-(((((((((((....)))))))))).(.(((((.((.(((........))).)).))))).).)))..)))...))))...))))))) ( -31.50) >consensus GGAA__AAGGGCGCUGGAAAAACAAGGUCGGACAAUUAGUCAGGUAAUUGACUGAUUAGAAAGCAAUGCCACAAAUAAAA__GC_AAUGUUU___G__AUGUUUAAUUUUCAUUUAAGCG .........((((((((..........)))).)((((((((((....))))))))))..........))).................................................. (-12.46 = -12.70 + 0.24)

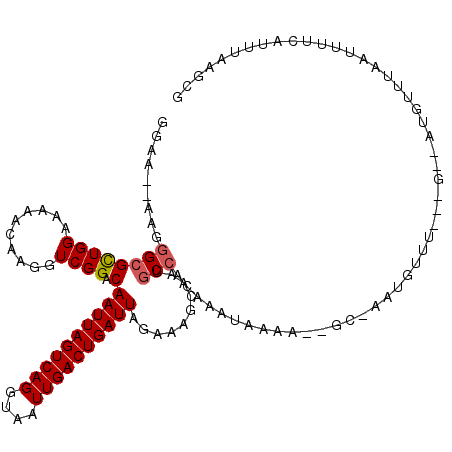
| Location | 8,862,024 – 8,862,132 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.51 |
| Mean single sequence MFE | -26.96 |
| Consensus MFE | -13.58 |
| Energy contribution | -13.94 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8862024 108 - 20766785 -----GCAAGCUAGUUUCCCGUGAAAAGCGGGUU---GGAAA-AAGGGCGCUGGAAAAACAAGGUCGGACAAUUAGUCAGGUAAUUGACUGAUUAGAAAGCAAUGCCAGAAAUAAAA -----(((.(((.(((((((((.....)))))..---.....-...(((..((......))..)))))))((((((((((....))))))))))....)))..)))........... ( -25.80) >DroSec_CAF1 106887 106 - 1 -----GCAAGCAAGUCGCCCGGAA-AAGGGGGUU---GGAA--AAGGGCGCUGGAAAAACAAGGUCGGACAAUUAGUCAGGUAAUUGACUGAUUAGAAAGCAAUGCCACAAAUAAAA -----(((.((..(((((((....-....)))).---....--...(((..((......))..))).)))((((((((((....)))))))))).....))..)))........... ( -25.10) >DroSim_CAF1 106798 107 - 1 -----GCAAGCAAGUUCCGCGGAAAAAGGGGGUU---GGAA--AAGGGCGCUGGAAAAACAAGGUCGGACAAUUAGUCAGGUAAUUGACUGAUUAGAAAGCAAUGCCACAAAUGAAA -----.....(((.((((.........)))).))---)...--...((((.((.........(......)((((((((((....))))))))))......)).)))).......... ( -21.50) >DroEre_CAF1 108569 95 - 1 GCAAUGCAAGCAAGUG-CUCGGAA-AAG-------------------GGGUUGGAAAAACCAGCUCGG-CAAUUAGUCAGGUAAUUGACUGAUUGCAAAGCAAUGCCACAAAUUAAA (((.(((.(((....)-)).....-...-------------------(((((((.....))))))).(-(((((((((((....))))))))))))...))).)))........... ( -34.10) >DroYak_CAF1 111009 110 - 1 -----GCAAGCAAGUCCCUCGGAA-AAGCGGGCUGUCGGAAAAAGGGGCGCUGGAAAAACAAGCUCGG-CAAUUAGUCAGGUAAUUGACUGAUUAGAAAGCAAUGCCACAAAUAAAA -----(((.((..(((((((((..-.((....)).))).....))))))(((((..........))))-)((((((((((....)))))))))).....))..)))........... ( -28.30) >consensus _____GCAAGCAAGUCCCCCGGAA_AAGCGGGUU___GGAA__AAGGGCGCUGGAAAAACAAGGUCGGACAAUUAGUCAGGUAAUUGACUGAUUAGAAAGCAAUGCCACAAAUAAAA .....(((.((...................................(((..((......))..)))....((((((((((....)))))))))).....))..)))........... (-13.58 = -13.94 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:50 2006