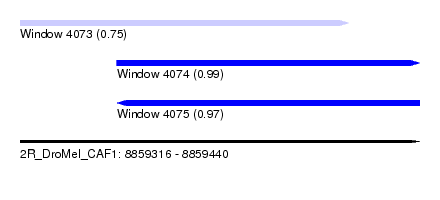
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,859,316 – 8,859,440 |
| Length | 124 |
| Max. P | 0.990397 |

| Location | 8,859,316 – 8,859,418 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -23.22 |
| Consensus MFE | -16.77 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8859316 102 + 20766785 AGCACA--CACAUACACACGCUCAAAGAAUGUGUAAGUGCAAAAGGCAUUUAUU----UCUGGACCCAAAUGAAGGCACGACCAGGCGUGGCUUCAAAUUGUAAUGAU .((...--(((.((((((..(.....)..)))))).)))......)).......----............(((((.((((......)))).)))))............ ( -24.00) >DroSec_CAF1 104200 79 + 1 AGC-------------------------AUGUGUAAGUGCAAAAGGCAUUUAUU----UCUGGACCCAAAUGAAGGCACGACCAGGCGUGGCUUCAAAUUGUAAUGAU ..(-------------------------(...((((((((.....)))))))).----..))........(((((.((((......)))).)))))............ ( -20.60) >DroSim_CAF1 104116 79 + 1 AGC-------------------------AUGUGUAAGUGCAAAAGGCAUUUAUU----UCUGGACCCAAAUGAAGGCACGACCAGGCGUGGCUUCAAAUUGUAAUGAU ..(-------------------------(...((((((((.....)))))))).----..))........(((((.((((......)))).)))))............ ( -20.60) >DroEre_CAF1 105864 99 + 1 -----ACACACACACACGCACUCAAAGGCUGUGUAAGUGCAAAAGGCAUUUAUA----UCUGGACCCAAAUGAUGGCACGACCAGGCGUGGCUUCAAAUUGUAAUGAU -----.........(((((...........((((((((((.....)))))))))----)((((..(((.....))).....))))))))).................. ( -23.40) >DroYak_CAF1 108186 104 + 1 GGCACACAAACACACACUCACUCAAAGGCUGUGUAAGUGCAAAAGGCAUUUAUU----UCUGGACCCAAAUGAUGGCACGACCAGGCGUGGCUUCAAAUUGUAAUGAU (((.(((..(((((..((.......))..)))))((((((.....))))))...----(((((..(((.....))).....))))).))))))............... ( -20.80) >DroPer_CAF1 119690 94 + 1 GGCAUUCAAAUAC--UC------------CCUGUAAGUGCAAAAGGCAUUUAUCCGGACCCGGACCUCAUUGCAGGCACUGCCAGGCGUGGCUCCGGAUUGUAAUGAU ..(((((((....--((------------(..((((((((.....))))))))..))).(((((((....(((.(((...)))..))).)).))))).))).)))).. ( -29.90) >consensus AGC__A___ACAC__AC___________AUGUGUAAGUGCAAAAGGCAUUUAUU____UCUGGACCCAAAUGAAGGCACGACCAGGCGUGGCUUCAAAUUGUAAUGAU ................................((((((((.....)))))))).................(((((.((((......)))).)))))............ (-16.77 = -17.30 + 0.53)

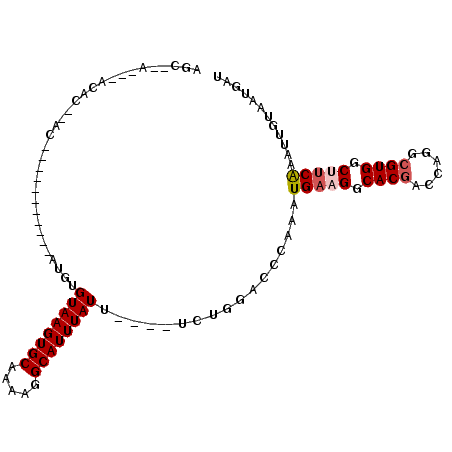
| Location | 8,859,346 – 8,859,440 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 87.80 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -27.69 |
| Energy contribution | -28.05 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8859346 94 + 20766785 GUAAGUGCAAAAGGCAUUUAUUUCUGGACCCAAAUGAAGGCACGACCAGGCGUGGCUUCAAAUUGUAAUGAUGCCACGCCCCC----------------ACACCCAUAGC ((((((((.....))))))))...(((...........((.....)).((((((((.(((........))).))))))))...----------------....))).... ( -28.80) >DroSec_CAF1 104207 93 + 1 GUAAGUGCAAAAGGCAUUUAUUUCUGGACCCAAAUGAAGGCACGACCAGGCGUGGCUUCAAAUUGUAAUGAUGCCACGCC-CC----------------ACACCCAACGC ((((((((.....))))))))...(((...........((.....)).((((((((.(((........))).))))))))-..----------------....))).... ( -29.00) >DroSim_CAF1 104123 94 + 1 GUAAGUGCAAAAGGCAUUUAUUUCUGGACCCAAAUGAAGGCACGACCAGGCGUGGCUUCAAAUUGUAAUGAUGCCACGCCCCC----------------ACACCCAACGC ((((((((.....))))))))...(((...........((.....)).((((((((.(((........))).))))))))...----------------....))).... ( -29.00) >DroEre_CAF1 105891 110 + 1 GUAAGUGCAAAAGGCAUUUAUAUCUGGACCCAAAUGAUGGCACGACCAGGCGUGGCUUCAAAUUGUAAUGAUGCCACGCCCCCACGGCCCCACUCCCCCACGCCCAUGGC ((((((((.....))))))))...(((..((......(((.....)))((((((((.(((........))).)))))))).....))..))).........(((...))) ( -34.40) >DroYak_CAF1 108218 101 + 1 GUAAGUGCAAAAGGCAUUUAUUUCUGGACCCAAAUGAUGGCACGACCAGGCGUGGCUUCAAAUUGUAAUGAUGCCACGCCCCCACG---------CCCCACGCCCAUCGC ((((((((.....))))))))..............(((((........((((((((.(((........))).)))))))).....(---------(.....))))))).. ( -32.20) >DroAna_CAF1 103701 93 + 1 -U-UAUGCUAAAGGCAUUUAUUUCUGGACCCAAAUGAAGGCACGACCAGGCGUGGCUUCAAAUUGUAAUGAUGCCACGCCCCCUC---------------CACCCACCAC -.-.((((.....)))).......((((..........((.....)).((((((((.(((........))).))))))))...))---------------))........ ( -25.90) >consensus GUAAGUGCAAAAGGCAUUUAUUUCUGGACCCAAAUGAAGGCACGACCAGGCGUGGCUUCAAAUUGUAAUGAUGCCACGCCCCC________________ACACCCAUCGC ((((((((.....))))))))...(((...........((.....)).((((((((.(((........))).)))))))).......................))).... (-27.69 = -28.05 + 0.36)

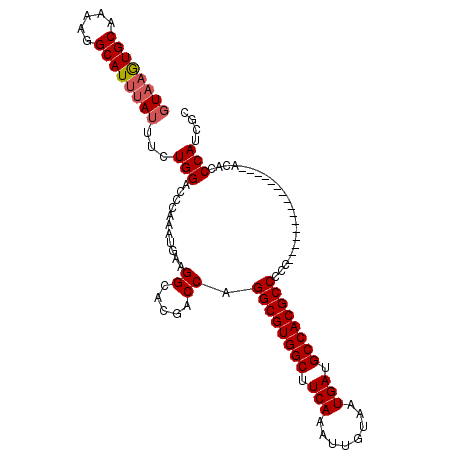
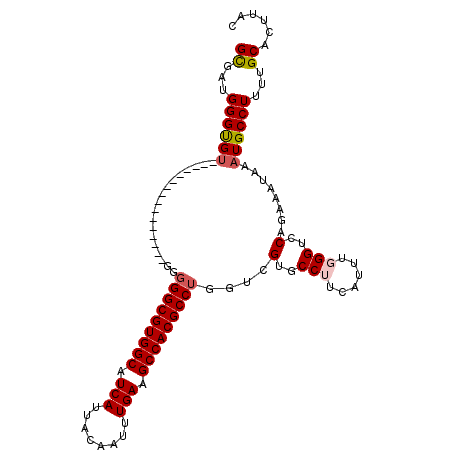
| Location | 8,859,346 – 8,859,440 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 87.80 |
| Mean single sequence MFE | -38.97 |
| Consensus MFE | -32.74 |
| Energy contribution | -33.05 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8859346 94 - 20766785 GCUAUGGGUGU----------------GGGGGCGUGGCAUCAUUACAAUUUGAAGCCACGCCUGGUCGUGCCUUCAUUUGGGUCCAGAAAUAAAUGCCUUUUGCACUUAC ((...((((((----------------..(((((((((.(((........))).)))))))))....(..(((......)))..)........))))))...))...... ( -35.10) >DroSec_CAF1 104207 93 - 1 GCGUUGGGUGU----------------GG-GGCGUGGCAUCAUUACAAUUUGAAGCCACGCCUGGUCGUGCCUUCAUUUGGGUCCAGAAAUAAAUGCCUUUUGCACUUAC (((..((((((----------------.(-((((((((.(((........))).)))))))))....(..(((......)))..)........))))))..)))...... ( -37.00) >DroSim_CAF1 104123 94 - 1 GCGUUGGGUGU----------------GGGGGCGUGGCAUCAUUACAAUUUGAAGCCACGCCUGGUCGUGCCUUCAUUUGGGUCCAGAAAUAAAUGCCUUUUGCACUUAC (((..((((((----------------..(((((((((.(((........))).)))))))))....(..(((......)))..)........))))))..)))...... ( -37.00) >DroEre_CAF1 105891 110 - 1 GCCAUGGGCGUGGGGGAGUGGGGCCGUGGGGGCGUGGCAUCAUUACAAUUUGAAGCCACGCCUGGUCGUGCCAUCAUUUGGGUCCAGAUAUAAAUGCCUUUUGCACUUAC ((...((((((.((((((((((((((...(((((((((.(((........))).)))))))))...)).))).))))))...)))........))))))...))...... ( -44.30) >DroYak_CAF1 108218 101 - 1 GCGAUGGGCGUGGGG---------CGUGGGGGCGUGGCAUCAUUACAAUUUGAAGCCACGCCUGGUCGUGCCAUCAUUUGGGUCCAGAAAUAAAUGCCUUUUGCACUUAC ((((.((((((((.(---------((...(((((((((.(((........))).)))))))))...))).))))..((((....)))).......)))).))))...... ( -45.60) >DroAna_CAF1 103701 93 - 1 GUGGUGGGUG---------------GAGGGGGCGUGGCAUCAUUACAAUUUGAAGCCACGCCUGGUCGUGCCUUCAUUUGGGUCCAGAAAUAAAUGCCUUUAGCAUA-A- ((...((((.---------------((..(((((((((.(((........))).)))))))))..))(..(((......)))..)..........))))...))...-.- ( -34.80) >consensus GCGAUGGGUGU________________GGGGGCGUGGCAUCAUUACAAUUUGAAGCCACGCCUGGUCGUGCCUUCAUUUGGGUCCAGAAAUAAAUGCCUUUUGCACUUAC ((...((((((..................(((((((((.(((........))).)))))))))....(..(((......)))..)........))))))...))...... (-32.74 = -33.05 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:47 2006