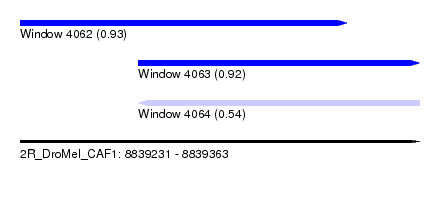
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,839,231 – 8,839,363 |
| Length | 132 |
| Max. P | 0.928982 |

| Location | 8,839,231 – 8,839,339 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 92.57 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -26.40 |
| Energy contribution | -26.48 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
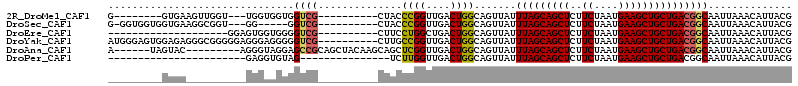
>2R_DroMel_CAF1 8839231 108 + 20766785 GACAAUUGCAUUAAAUUCGCCUGCACAACGGUGCGUAUGCGUAAUGUUUAAUUGCCGUCAGCAGCUUCAUUAGAAGAGCUGCUAAAUAACUGCCAGUCAACCGGGUAG----- (((....(((((((((.(((.(((((....)))))...)))....)))))).))).)))((((((((........))))))))......(((((.........)))))----- ( -31.90) >DroPse_CAF1 94376 106 + 1 GACAAAUGCAUUAAAUUUUCCUGCACAGCAGUGCGUAUGCGUAAUGUUUAAUUGCCGUCAGCAGCUUCAUUAGAAGAGCUGCUAAAUAACUGCCAGUCAACCAAGA------- (((...((((...........))))..(((((..(.(((.(((((.....)))))))))((((((((........)))))))).....)))))..)))........------- ( -27.70) >DroEre_CAF1 85949 108 + 1 GACAAUUGCAUUAAAUUCGCCUGCACAACGGUGCGUAUGCGUAAUGUUUAAUUGCCGUCAGCAGCUUCAUUAGAAGAGCUGCUAAAUAACUGCCAGUCAGCCAGGAAG----- (((....(((((((((.(((.(((((....)))))...)))....)))))).))).)))((((((((........))))))))......(((.(.....).)))....----- ( -29.00) >DroYak_CAF1 86967 108 + 1 GACAAUUGCAUUAAAUUCGCCUGCACAACGGUGCGUAUGCGUAAUGUUUAAUUGCCGUCAGCAGCUUCAUUAGAAGAGCUGCUAAAUAACUGCCAGUCAACCGGCAAG----- (((....(((((((((.(((.(((((....)))))...)))....)))))).))).)))((((((((........)))))))).......((((........))))..----- ( -32.60) >DroAna_CAF1 84472 113 + 1 GACAAUUGCAUUAAAUUUGCCUGCACAACGGUGCGUAUGCGUAAUGUUUAAUUGCCGUCAGCAGCUUCAUUAGAAGAGCUGCUAAAUAACUGCCAGUCAACCGAGCUGCUUGU (((....(((((((((.(((.(((((....)))))...)))....)))))).))).)))((((((((........))))))))........((.(((.......))))).... ( -27.60) >DroPer_CAF1 97484 106 + 1 GACAAAUGCAUUAAAUUUUCCUGCACAGCAGUGCGUAUGCGUAAUGUUUAAUUGCCGUCAGCAGCUUCAUUAGAAGAGCUGCUAAAUAACUGCCAGUCAACCAAGA------- (((...((((...........))))..(((((..(.(((.(((((.....)))))))))((((((((........)))))))).....)))))..)))........------- ( -27.70) >consensus GACAAUUGCAUUAAAUUCGCCUGCACAACGGUGCGUAUGCGUAAUGUUUAAUUGCCGUCAGCAGCUUCAUUAGAAGAGCUGCUAAAUAACUGCCAGUCAACCAAGAAG_____ (((....(((((((((.(((.(((((....)))))...)))....)))))).)))....((((((((........))))))))............)))............... (-26.40 = -26.48 + 0.08)

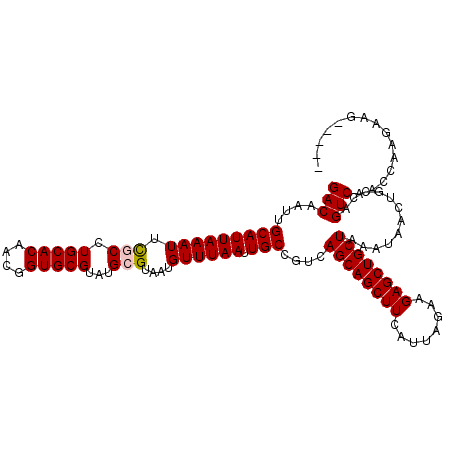
| Location | 8,839,270 – 8,839,363 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 73.41 |
| Mean single sequence MFE | -21.52 |
| Consensus MFE | -14.00 |
| Energy contribution | -14.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8839270 93 + 20766785 CGUAAUGUUUAAUUGCCGUCAGCAGCUUCAUUAGAAGAGCUGCUAAAUAACUGCCAGUCAACCGGGUAG----------CGACCACCACCA---ACCAACUUCAC--------C .(((((.....))))).(((((((((((........))))))))......(((((.........)))))----------.)))........---...........--------. ( -21.90) >DroSec_CAF1 84091 95 + 1 CGUAAUGUUUAAUUGCCGUCAGCAGCUUCAUUAGAAGAGCUGCUAAAUAACUGCCAGUCAACCGGGUAG----------CGACC-----CC---ACCGCCUUCACCACCACC-C .(((((.....))))).(((((((((((........))))))))......(((((.........)))))----------.))).-----..---..................-. ( -21.90) >DroEre_CAF1 85988 84 + 1 CGUAAUGUUUAAUUGCCGUCAGCAGCUUCAUUAGAAGAGCUGCUAAAUAACUGCCAGUCAGCCAGGAAG----------CGACCCCACCACUCC-------------------- .(((((.....))))).(((((((((((........))))))))......(((.(.....).)))....----------.)))...........-------------------- ( -18.60) >DroYak_CAF1 87006 104 + 1 CGUAAUGUUUAAUUGCCGUCAGCAGCUUCAUUAGAAGAGCUGCUAAAUAACUGCCAGUCAACCGGCAAG----------CGACCCCCUCCCUCCCCCGCCCUCUCCACUCCCAU .(((((.....))))).(((((((((((........)))))))).......((((........))))..----------.)))............................... ( -22.20) >DroAna_CAF1 84511 99 + 1 CGUAAUGUUUAAUUGCCGUCAGCAGCUUCAUUAGAAGAGCUGCUAAAUAACUGCCAGUCAACCGAGCUGCUUGUAGCUGCGGCUCCUACCCU---------GUACUA------U .(((((.....)))))....((((((((........))))))))..........(((......(((((((........))))))).....))---------).....------. ( -27.20) >DroPer_CAF1 97523 76 + 1 CGUAAUGUUUAAUUGCCGUCAGCAGCUUCAUUAGAAGAGCUGCUAAAUAACUGCCAGUCAACCAAGA---------------CUACACCUC----------------------- .(((((.....)))))....((((((((........))))))))...........((((......))---------------)).......----------------------- ( -17.30) >consensus CGUAAUGUUUAAUUGCCGUCAGCAGCUUCAUUAGAAGAGCUGCUAAAUAACUGCCAGUCAACCGGGAAG__________CGACCCCAACCC__________UCAC_________ .(((((.....)))))....((((((((........))))))))...................................................................... (-14.00 = -14.00 + 0.00)

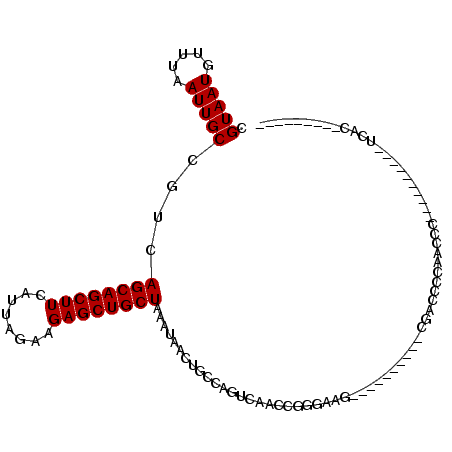
| Location | 8,839,270 – 8,839,363 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 73.41 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -18.02 |
| Energy contribution | -17.97 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
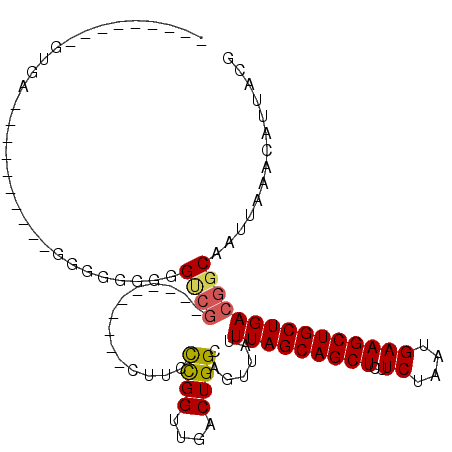
>2R_DroMel_CAF1 8839270 93 - 20766785 G--------GUGAAGUUGGU---UGGUGGUGGUCG----------CUACCCGGUUGACUGGCAGUUAUUUAGCAGCUCUUCUAAUGAAGCUGCUGACGGCAAUUAAACAUUACG .--------((((.((..((---(((((((....)----------))))).....)))..)).(((..(((((((((..((....))))))))))).))).........)))). ( -29.50) >DroSec_CAF1 84091 95 - 1 G-GGUGGUGGUGAAGGCGGU---GG-----GGUCG----------CUACCCGGUUGACUGGCAGUUAUUUAGCAGCUCUUCUAAUGAAGCUGCUGACGGCAAUUAAACAUUACG .-.((((((.((..(((..(---.(-----(((..----------..)))).)..).))..))(((..(((((((((..((....))))))))))).))).......)))))). ( -30.20) >DroEre_CAF1 85988 84 - 1 --------------------GGAGUGGUGGGGUCG----------CUUCCUGGCUGACUGGCAGUUAUUUAGCAGCUCUUCUAAUGAAGCUGCUGACGGCAAUUAAACAUUACG --------------------...((((((.(((((----------((....))).)))).((.(((....(((((((..((....)))))))))))).)).......)))))). ( -26.70) >DroYak_CAF1 87006 104 - 1 AUGGGAGUGGAGAGGGCGGGGGAGGGAGGGGGUCG----------CUUGCCGGUUGACUGGCAGUUAUUUAGCAGCUCUUCUAAUGAAGCUGCUGACGGCAAUUAAACAUUACG .....((((..((..((.(..(((.((.....)).----------)))(((((....)))))......(((((((((..((....)))))))))))).))..))...))))... ( -29.50) >DroAna_CAF1 84511 99 - 1 A------UAGUAC---------AGGGUAGGAGCCGCAGCUACAAGCAGCUCGGUUGACUGGCAGUUAUUUAGCAGCUCUUCUAAUGAAGCUGCUGACGGCAAUUAAACAUUACG .------..(((.---------.(..(((..((((..((.....)).((.(((....)))))......(((((((((..((....)))))))))))))))..)))..)..))). ( -29.00) >DroPer_CAF1 97523 76 - 1 -----------------------GAGGUGUAG---------------UCUUGGUUGACUGGCAGUUAUUUAGCAGCUCUUCUAAUGAAGCUGCUGACGGCAAUUAAACAUUACG -----------------------.....((((---------------(.(((((((.(((........(((((((((..((....)))))))))))))))))))))..))))). ( -20.70) >consensus _________GUGA__________GGGGGGGGGUCG__________CUUCCCGGUUGACUGGCAGUUAUUUAGCAGCUCUUCUAAUGAAGCUGCUGACGGCAAUUAAACAUUACG ...............................((((..............((((....)))).......(((((((((..((....))))))))))))))).............. (-18.02 = -17.97 + -0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:37 2006