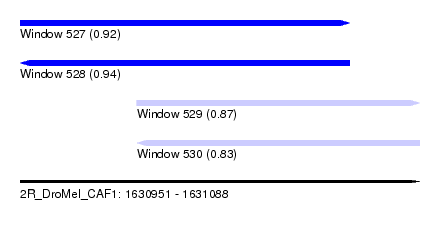
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,630,951 – 1,631,088 |
| Length | 137 |
| Max. P | 0.940216 |

| Location | 1,630,951 – 1,631,064 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.70 |
| Mean single sequence MFE | -43.87 |
| Consensus MFE | -29.02 |
| Energy contribution | -30.30 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
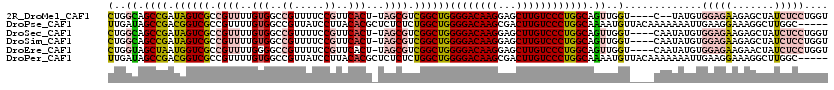
>2R_DroMel_CAF1 1630951 113 + 20766785 CUGGCAGCCGAUAGUCGCCGUUUUGUGGCCGUUUUCCGUUCACU-UAGCGUCGGCUGGGGACAAGGAGCUUGUCCCUGGCAGUUGGU----C--UAUGUGGAGAAGAGCUAUCUCCUGGU (..((.((((.((((((.((((..(((..((.....))..))).-.)))).))))))((((((((...)))))))))))).))..).----.--.....(((((.......))))).... ( -45.50) >DroPse_CAF1 42527 115 + 1 UUGAUAGCCGACGGUCGCCGUUUUGUGGCCGUUAUCCUUACACGCUCUCUCUGGCUGGGGACAAGCGACUUGUCCCUGGCAAAAUGUUACAAAAAAAUUGAAGGAAAGGCUUGGC----- .....(((((((((((((......))))))))).(((((..............((..((((((((...))))))))..)).........(((.....))))))))..))))....----- ( -43.40) >DroSec_CAF1 16767 115 + 1 CUGGCAGCCGAUAGUCGCCGUUUUGUGGCCGUUUUCCGUUCACU-UAGCGUCGGCUGGGGACAAGGAGCUUGUCCCUGGCAGUUGGU----CAAUAUGUGGAGAAGAGCUAUCUCCUGGU (..((.((((.((((((.((((..(((..((.....))..))).-.)))).))))))((((((((...)))))))))))).))..).----........(((((.......))))).... ( -45.50) >DroSim_CAF1 15204 115 + 1 CUGGCAGCCGAUAGUCGCCGUUUUGUGGCCGUUUUCCGUUCACU-UAGCGUCGGCUGGGGACAAGGAGCUUGUCCCUGGCAGUUGGU----CAAUAUGUGGAGAAGAGCUAUCUCCUGGU (..((.((((.((((((.((((..(((..((.....))..))).-.)))).))))))((((((((...)))))))))))).))..).----........(((((.......))))).... ( -45.50) >DroEre_CAF1 19267 115 + 1 CUGGUAGCUAAUGGUCGCCGUUUUGGGGCCGUUUUCCGUUCACU-UAGCGUCGGCUGGGGACAAGGAGCUUGUCCCUGGCAGUUGGU----CAAUAUGUGGAGAAGAACUAUCUCCUGGU ..((.....(((((((.((.....)))))))))..))(((.(((-.(((....((..((((((((...))))))))..)).))))))----.)))....(((((.......))))).... ( -39.90) >DroPer_CAF1 37083 115 + 1 UUGAUAGCCGACGGUCGCCGUUUUGUGGCCGUUAUCCUUACACGCUCUCUCUGGCUGGGGACAAGCGACUUGUCCCUGGCAAAAUGUUACAAAAAAAUUGAAGGAAAGGCUUGGC----- .....(((((((((((((......))))))))).(((((..............((..((((((((...))))))))..)).........(((.....))))))))..))))....----- ( -43.40) >consensus CUGGCAGCCGAUAGUCGCCGUUUUGUGGCCGUUUUCCGUUCACU_UAGCGUCGGCUGGGGACAAGGAGCUUGUCCCUGGCAGUUGGU____CAAUAUGUGGAGAAGAGCUAUCUCCUGGU (..((.((((.((((((.((((..(((..((.....))..)))...)))).))))))((((((((...)))))))))))).))..).............(((((.......))))).... (-29.02 = -30.30 + 1.28)
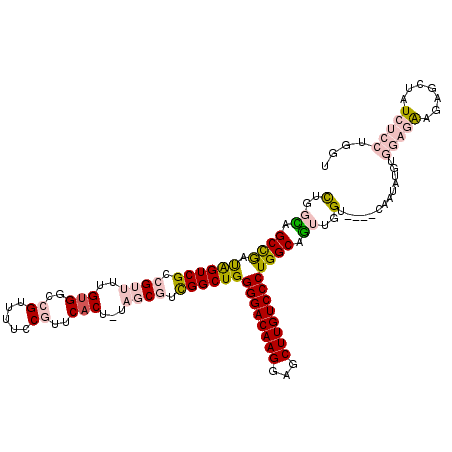
| Location | 1,630,951 – 1,631,064 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.70 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -23.70 |
| Energy contribution | -24.57 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1630951 113 - 20766785 ACCAGGAGAUAGCUCUUCUCCACAUA--G----ACCAACUGCCAGGGACAAGCUCCUUGUCCCCAGCCGACGCUA-AGUGAACGGAAAACGGCCACAAAACGGCGACUAUCGGCUGCCAG ....(((((.......))))).....--.----.....(((...((((((((...))))))))(((((((((((.-.(((..((.....))..))).....))))....))))))).))) ( -39.80) >DroPse_CAF1 42527 115 - 1 -----GCCAAGCCUUUCCUUCAAUUUUUUUGUAACAUUUUGCCAGGGACAAGUCGCUUGUCCCCAGCCAGAGAGAGCGUGUAAGGAUAACGGCCACAAAACGGCGACCGUCGGCUAUCAA -----(((..(((..(((((((.((((((((.........((..((((((((...))))))))..))))))))))...)).)))))....)))........(((....))))))...... ( -35.80) >DroSec_CAF1 16767 115 - 1 ACCAGGAGAUAGCUCUUCUCCACAUAUUG----ACCAACUGCCAGGGACAAGCUCCUUGUCCCCAGCCGACGCUA-AGUGAACGGAAAACGGCCACAAAACGGCGACUAUCGGCUGCCAG ....(((((.......)))))........----.....(((...((((((((...))))))))(((((((((((.-.(((..((.....))..))).....))))....))))))).))) ( -39.80) >DroSim_CAF1 15204 115 - 1 ACCAGGAGAUAGCUCUUCUCCACAUAUUG----ACCAACUGCCAGGGACAAGCUCCUUGUCCCCAGCCGACGCUA-AGUGAACGGAAAACGGCCACAAAACGGCGACUAUCGGCUGCCAG ....(((((.......)))))........----.....(((...((((((((...))))))))(((((((((((.-.(((..((.....))..))).....))))....))))))).))) ( -39.80) >DroEre_CAF1 19267 115 - 1 ACCAGGAGAUAGUUCUUCUCCACAUAUUG----ACCAACUGCCAGGGACAAGCUCCUUGUCCCCAGCCGACGCUA-AGUGAACGGAAAACGGCCCCAAAACGGCGACCAUUAGCUACCAG ....(((((.......)))))........----.....(.((..((((((((...))))))))..)).)..((((-(.((..((.....))(((.......)))...)))))))...... ( -31.20) >DroPer_CAF1 37083 115 - 1 -----GCCAAGCCUUUCCUUCAAUUUUUUUGUAACAUUUUGCCAGGGACAAGUCGCUUGUCCCCAGCCAGAGAGAGCGUGUAAGGAUAACGGCCACAAAACGGCGACCGUCGGCUAUCAA -----(((..(((..(((((((.((((((((.........((..((((((((...))))))))..))))))))))...)).)))))....)))........(((....))))))...... ( -35.80) >consensus ACCAGGAGAUAGCUCUUCUCCACAUAUUG____ACCAACUGCCAGGGACAAGCUCCUUGUCCCCAGCCGACGCUA_AGUGAACGGAAAACGGCCACAAAACGGCGACCAUCGGCUACCAG ....(((((.......))))).................(((...((((((((...)))))))).((((((.......((.........)).(((.......))).....))))))..))) (-23.70 = -24.57 + 0.86)

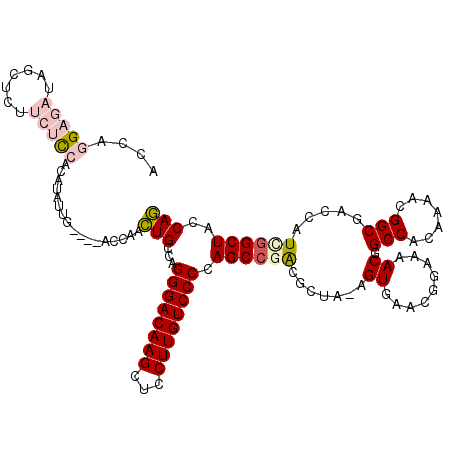
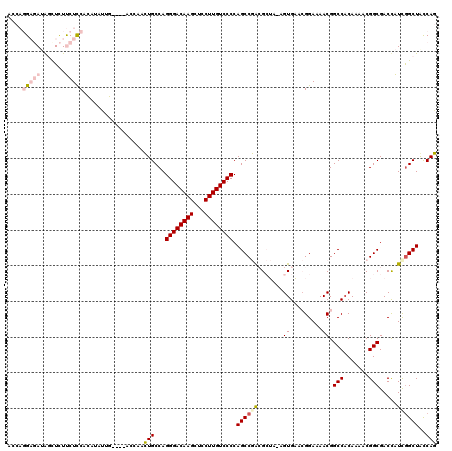
| Location | 1,630,991 – 1,631,088 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 95.96 |
| Mean single sequence MFE | -36.58 |
| Consensus MFE | -32.54 |
| Energy contribution | -33.10 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1630991 97 + 20766785 CACUUAGCGUCGGCUGGGGACAAGGAGCUUGUCCCUGGCAGUUGGUC--UAUGUGGAGAAGAGCUAUCUCCUGGUAAGAUCCUCUGUUGCUUUACGAAG .....((((.(.((..((((((((...))))))))..))((..((((--(((..(((((.......)))))..)).)))))..))).))))........ ( -34.40) >DroSec_CAF1 16807 99 + 1 CACUUAGCGUCGGCUGGGGACAAGGAGCUUGUCCCUGGCAGUUGGUCAAUAUGUGGAGAAGAGCUAUCUCCUGGUAAGAUCCUCUGUUGCUGUGUGAAU (((.(((((.(.((..((((((((...))))))))..))((..((((..(((..(((((.......)))))..))).))))..))).))))).)))... ( -38.40) >DroSim_CAF1 15244 99 + 1 CACUUAGCGUCGGCUGGGGACAAGGAGCUUGUCCCUGGCAGUUGGUCAAUAUGUGGAGAAGAGCUAUCUCCUGGUAAGAUCCUCUGUUGCAGUGCGAAG ((((((((....((..((((((((...))))))))..))((..((((..(((..(((((.......)))))..))).))))..)))))).))))..... ( -35.70) >DroEre_CAF1 19307 99 + 1 CACUUAGCGUCGGCUGGGGACAAGGAGCUUGUCCCUGGCAGUUGGUCAAUAUGUGGAGAAGAACUAUCUCCUGGUAAGAUCCUCUGUUGCUGUGCGAAG ..(((.(((.((((.(((((((((...)))))))))(((((..((((..(((..(((((.......)))))..))).))))..)))))))))))).))) ( -37.80) >consensus CACUUAGCGUCGGCUGGGGACAAGGAGCUUGUCCCUGGCAGUUGGUCAAUAUGUGGAGAAGAGCUAUCUCCUGGUAAGAUCCUCUGUUGCUGUGCGAAG ......(((.((((.(((((((((...)))))))))(((((..((((..(((..(((((.......)))))..))).))))..)))))))))))).... (-32.54 = -33.10 + 0.56)

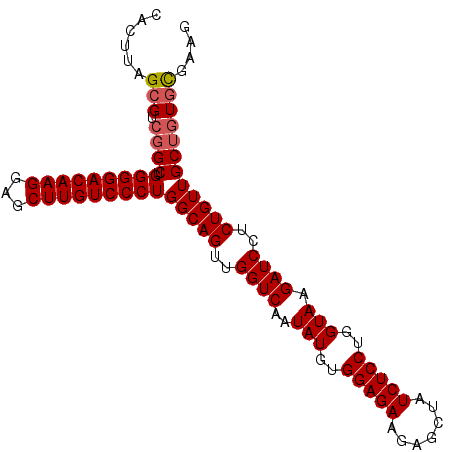
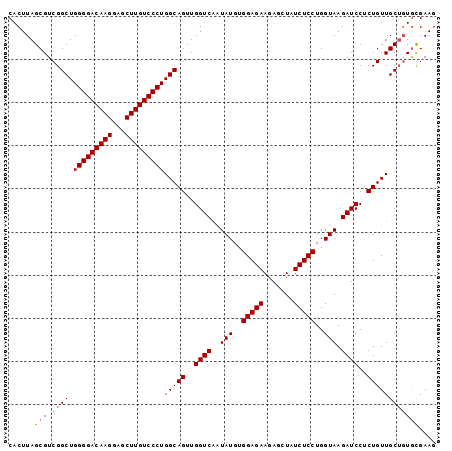
| Location | 1,630,991 – 1,631,088 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 95.96 |
| Mean single sequence MFE | -27.79 |
| Consensus MFE | -25.84 |
| Energy contribution | -25.72 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1630991 97 - 20766785 CUUCGUAAAGCAACAGAGGAUCUUACCAGGAGAUAGCUCUUCUCCACAUA--GACCAACUGCCAGGGACAAGCUCCUUGUCCCCAGCCGACGCUAAGUG ........(((......((.(((.....(((((.......)))))....)--))))..(.((..((((((((...))))))))..)).)..)))..... ( -28.10) >DroSec_CAF1 16807 99 - 1 AUUCACACAGCAACAGAGGAUCUUACCAGGAGAUAGCUCUUCUCCACAUAUUGACCAACUGCCAGGGACAAGCUCCUUGUCCCCAGCCGACGCUAAGUG ...(((..(((......((.((......(((((.......))))).......))))..(.((..((((((((...))))))))..)).)..)))..))) ( -28.22) >DroSim_CAF1 15244 99 - 1 CUUCGCACUGCAACAGAGGAUCUUACCAGGAGAUAGCUCUUCUCCACAUAUUGACCAACUGCCAGGGACAAGCUCCUUGUCCCCAGCCGACGCUAAGUG .....((((((......((.((......(((((.......))))).......))))..(.((..((((((((...))))))))..)).)..))..)))) ( -27.02) >DroEre_CAF1 19307 99 - 1 CUUCGCACAGCAACAGAGGAUCUUACCAGGAGAUAGUUCUUCUCCACAUAUUGACCAACUGCCAGGGACAAGCUCCUUGUCCCCAGCCGACGCUAAGUG ...(((..(((......((.((......(((((.......))))).......))))..(.((..((((((((...))))))))..)).)..)))..))) ( -27.82) >consensus CUUCGCACAGCAACAGAGGAUCUUACCAGGAGAUAGCUCUUCUCCACAUAUUGACCAACUGCCAGGGACAAGCUCCUUGUCCCCAGCCGACGCUAAGUG ...(((..(((......((.((......(((((.......))))).......))))..(.((..((((((((...))))))))..)).)..)))..))) (-25.84 = -25.72 + -0.12)

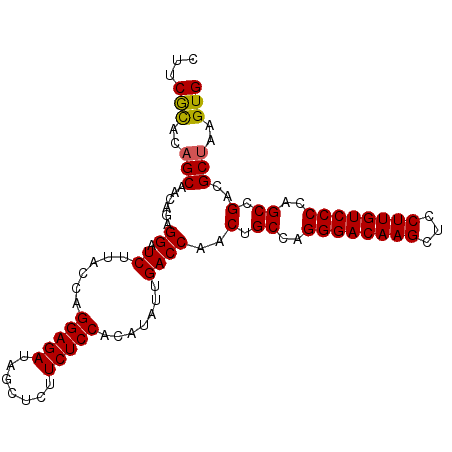
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:27 2006