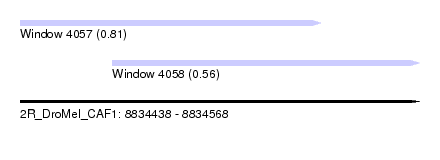
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,834,438 – 8,834,568 |
| Length | 130 |
| Max. P | 0.809492 |

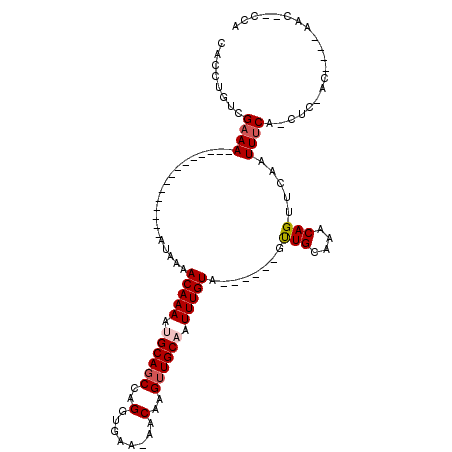
| Location | 8,834,438 – 8,834,536 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -20.98 |
| Consensus MFE | -13.69 |
| Energy contribution | -14.75 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8834438 98 + 20766785 -------CGGGCUG---CAUCAGCGUAAUUGCGGUUAUUUCACCUGUCGAAACAGAAAAUAAAAAAA--AAAACAAAACGCAGCCAGGUGAA-AACAAGUUGCAAUUUGUA------ -------..(((((---(....(((....)))..((((((...((((....)))).)))))).....--..........)))))).......-.((((((....)))))).------ ( -21.20) >DroPse_CAF1 88325 84 + 1 --------GGGCAG---AAUCAGCGUAAUUGCGGUUAUUUCACCUGUCGAAA---------------AUAAAACAAAAUGCAGCCAGGUGAA-AACAAGUUGCAAUUUGUA------ --------.(((((---.....(((....)))((.....))..)))))....---------------.....(((((.((((((...((...-.))..)))))).))))).------ ( -21.80) >DroGri_CAF1 92655 95 + 1 ------UGAGGCAACGACAUCAGCGUAAUUGCAGUUAUUUCACCUGUCGCAA---------------AUAAAACAAAAUGCAGCCGGGCGCA-AACAAGUUGCAAUUUGUACUCGCU ------.(((....(((((...(((....))).(......)...)))))...---------------.....(((((.((((((..(.....-..)..)))))).))))).)))... ( -21.10) >DroWil_CAF1 113795 85 + 1 -------AAGGGGG---UAGCCACGUAAUUGCGGUUAUUUCACCUGUCGAAA---------------AUAAAACAAAAUGCAUCGAGGCGAA-AACAAGUUGCAAUUUGUA------ -------...(..(---(((((.((....)).))))))..)...........---------------.....(((((.((((.(..(.....-..)..).)))).))))).------ ( -19.30) >DroAna_CAF1 79995 93 + 1 UAUUUUUGUGGCAG---CAUCAGCGUAAUUGCGGUUAUUCCACCUGUCGAAA---------------AUAAAACAAAACGCAGCCAGGUGAAAAACAAGUUGCAAUUUGUA------ (((((((..(((((---.....(((....)))((.....))..)))))))))---------------)))..(((((..(((((...((.....))..)))))..))))).------ ( -20.70) >DroPer_CAF1 91422 84 + 1 --------GGGCAG---AAUCAGCGUAAUUGCGGUUAUUUCACCUGUCGAAA---------------AUAAAACAAAAUGCAGCCAGGUGAA-AACAAGUUGCAAUUUGUA------ --------.(((((---.....(((....)))((.....))..)))))....---------------.....(((((.((((((...((...-.))..)))))).))))).------ ( -21.80) >consensus ________GGGCAG___CAUCAGCGUAAUUGCGGUUAUUUCACCUGUCGAAA_______________AUAAAACAAAAUGCAGCCAGGUGAA_AACAAGUUGCAAUUUGUA______ .........((((.........(((....)))(((......)))))))........................(((((.((((((..(........)..)))))).)))))....... (-13.69 = -14.75 + 1.06)

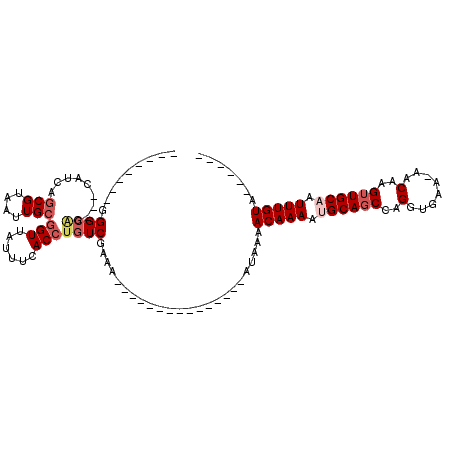
| Location | 8,834,468 – 8,834,568 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.49 |
| Mean single sequence MFE | -17.35 |
| Consensus MFE | -9.87 |
| Energy contribution | -10.32 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8834468 100 + 20766785 CACCUGUCGAAACAGAAAAUAAAAAAA--AAAACAAAACGCAGCCAGGUGAA-AACAAGUUGCAAUUUGUA------GUUGCAAACAGUUCAAUUUCA-CUC-GU----UAC--CCA ...((((....))))............--.......((((......((((((-(((...(((((((.....------)))))))...))....)))))-)))-))----)..--... ( -15.90) >DroPse_CAF1 88354 90 + 1 CACCUGUCGAAA---------------AUAAAACAAAAUGCAGCCAGGUGAA-AACAAGUUGCAAUUUGUA------GCUGCAAACAGUUCAAUUUCA-CUC-ACGUA-CAC--CCA ....(((((...---------------.....(((((.((((((...((...-.))..)))))).)))))(------((((....)))))........-...-.)).)-)).--... ( -18.90) >DroGri_CAF1 92689 95 + 1 CACCUGUCGCAA---------------AUAAAACAAAAUGCAGCCGGGCGCA-AACAAGUUGCAAUUUGUACUCGCUGUUGCAAACAGUUCAAUUUCUACUUUGU----UCC--ACA ....((..((((---------------(....(((((.((((((..(.....-..)..)))))).)))))....(((((.....)))))...........)))))----..)--).. ( -18.70) >DroWil_CAF1 113825 90 + 1 CACCUGUCGAAA---------------AUAAAACAAAAUGCAUCGAGGCGAA-AACAAGUUGCAAUUUGUA------GUUGCAAACAGUUCAAUUUCA-CG--ACACACAAC--CCG ....(((((...---------------............((......))(((-......(((((((.....------)))))))....))).......-))--)))......--... ( -15.70) >DroAna_CAF1 80032 90 + 1 CACCUGUCGAAA---------------AUAAAACAAAACGCAGCCAGGUGAAAAACAAGUUGCAAUUUGUA------GUUGCAAACAGUUCAAUUUCA-CUC-GC----AACUCCCA ............---------------...............((..((((((((((...(((((((.....------)))))))...)))...)))))-)).-))----........ ( -16.00) >DroPer_CAF1 91451 90 + 1 CACCUGUCGAAA---------------AUAAAACAAAAUGCAGCCAGGUGAA-AACAAGUUGCAAUUUGUA------GCUGCAAACAGUUCAAUUUCA-CUC-ACGUA-CAC--CCA ....(((((...---------------.....(((((.((((((...((...-.))..)))))).)))))(------((((....)))))........-...-.)).)-)).--... ( -18.90) >consensus CACCUGUCGAAA_______________AUAAAACAAAAUGCAGCCAGGUGAA_AACAAGUUGCAAUUUGUA______GUUGCAAACAGUUCAAUUUCA_CUC_AC____AAC__CCA ........((((....................(((((.((((((..(........)..)))))).)))))........(((....))).....)))).................... ( -9.87 = -10.32 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:31 2006