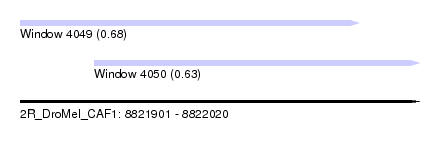
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,821,901 – 8,822,020 |
| Length | 119 |
| Max. P | 0.683742 |

| Location | 8,821,901 – 8,822,002 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -23.07 |
| Consensus MFE | -15.25 |
| Energy contribution | -14.70 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
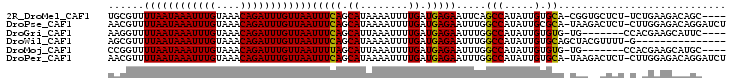
>2R_DroMel_CAF1 8821901 101 + 20766785 -----ACUGG-----AAACG--AGCGA-----GUCGCGC-UGCGUUUUAAUAAAUUUGUAAACAGAUUUGUUAAUUUCAGCAUAAAAUUUUGAUGAGAAUUCAGCCAUAUUGUGCA-CGG -----.((((-----(((((--((((.-----....)))-).))))))((((((((((....)))))))))).......((((((.((.((((.......))))..)).)))))).-))) ( -24.90) >DroPse_CAF1 74785 106 + 1 -----CCGAA-----AAAUG--AGCGAAGAACGGAGAGC-AACGUUUUAAUAAAUUUGUAAACAGAUUUGUUAAUUUCAGCAUAAAAUUUUGAUGAGAAUUUGGCCAUAUUGCGCA-UAA -----.....-----..(((--.((((((((((......-..))))((((((((((((....)))))))))))))))).((...(((((((....))))))).))......)).))-).. ( -21.50) >DroGri_CAF1 77741 106 + 1 GCAGGCAGUC-----AAAUG--AGCGA-----ACUGUGCGAAGGUUUUAAUAAAUUUGUAAACAGAUUUGUUAAUUUCAGCAUUAAAUUUUGAUGAGAAUUUGGCCAUAUUGUGUG-UG- (((.(((((.-----..(((--.((..-----...((((...((..((((((((((((....))))))))))))..)).))))((((((((....)))))))))))))))))).))-).- ( -27.80) >DroWil_CAF1 98712 105 + 1 -----CUGAA-----AAGUAGAAGCAAA----AAUGAGC-AGCGUUUUAAUAAAUUUGUAAACAGAUUUGUUAAUUUCAGCAUAAAAUUUUGAUGAGAAUUUGGCCAUAUUGUGCAGCUA -----.....-----...(((..(((..----.(((.((-.((...((((((((((((....)))))))))))).....))...(((((((....))))))).)))))....)))..))) ( -21.50) >DroAna_CAF1 66459 106 + 1 -----CCUGGCCCCCAAAUG--AGUGA-----GUCGAGG-AACGUUUUAAUAAAUUUGUAAACAGAUUUGUUAAUUUCAGCAUAAAAUUUUGAUGAGAAUUCAGCAAUAUUGUGCA-CUG -----((((((.(.(.....--.).).-----))).)))-...(..((((((((((((....))))))))))))...).((((((.(((((((.......)))).))).)))))).-... ( -22.00) >DroPer_CAF1 75534 106 + 1 -----CCGGA-----AAAUG--AGCGAAGAACGGAGAGC-AACGUUUUAAUAAAUUUGUAAACAGAUUUGUUAAUUUCAGCAUAAAAUUUUGAUGAGAAUUUGGCCAUAUUGUGCA-UAA -----.((.(-----(.(((--.((.(((((((......-..))))((((((((((((....))))))))))))(((((.((........)).))))).))).))))).)).))..-... ( -20.70) >consensus _____CCGGA_____AAAUG__AGCGA_____GGAGAGC_AACGUUUUAAUAAAUUUGUAAACAGAUUUGUUAAUUUCAGCAUAAAAUUUUGAUGAGAAUUUGGCCAUAUUGUGCA_CAA .......................(((....................((((((((((((....))))))))))))(((((.((........)).)))))..............)))..... (-15.25 = -14.70 + -0.55)

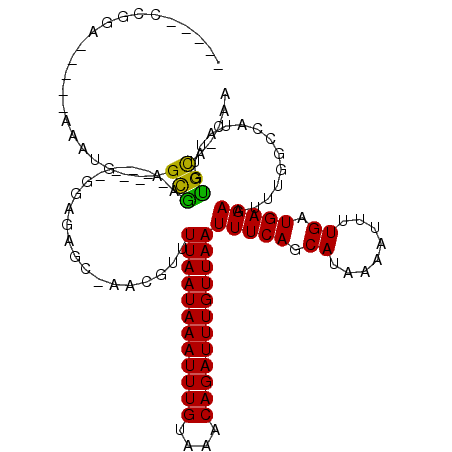
| Location | 8,821,923 – 8,822,020 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 78.50 |
| Mean single sequence MFE | -19.70 |
| Consensus MFE | -12.66 |
| Energy contribution | -12.30 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8821923 97 + 20766785 UGCGUUUUAAUAAAUUUGUAAACAGAUUUGUUAAUUUCAGCAUAAAAUUUUGAUGAGAAUUCAGCCAUAUUGUGCA-CGGUGCUCU-UCUGAAGACAGC---- ..(((.((((((((((((....)))))))))))).....((((((.((.((((.......))))..)).)))))))-))((..((.-...))..))...---- ( -17.80) >DroPse_CAF1 74812 101 + 1 AACGUUUUAAUAAAUUUGUAAACAGAUUUGUUAAUUUCAGCAUAAAAUUUUGAUGAGAAUUUGGCCAUAUUGCGCA-UAAGACUCU-CUUGGAGACAGGAUCU ..(((.((((((((((((....)))))))))))).....((...(((((((....))))))).))......)))..-........(-((((....)))))... ( -21.20) >DroGri_CAF1 77769 91 + 1 AAGGUUUUAAUAAAUUUGUAAACAGAUUUGUUAAUUUCAGCAUUAAAUUUUGAUGAGAAUUUGGCCAUAUUGUGUG-UG-------CCACGAAGCAUUC---- ..((..((((((((((((....))))))))))))..)).((...(((((((....)))))))((((((.....)))-.)-------)).....))....---- ( -21.60) >DroWil_CAF1 98737 87 + 1 AGCGUUUUAAUAAAUUUGUAAACAGAUUUGUUAAUUUCAGCAUAAAAUUUUGAUGAGAAUUUGGCCAUAUUGUGCAGCUACGUUUU-G--------------- ((((..((((((((((((....))))))))))))...).((((((((((((....))))))........)))))).))).......-.--------------- ( -17.00) >DroMoj_CAF1 81643 91 + 1 CCGGUUUUAAUAAAUUUGUAAACAGAUUUGUUAAUUUUAGCAUUAAAUUUUGAUGAGAAUUUGGCCAUAUUGUGUG-UG-------CCACGAAGCAUGC---- ..((..((((((((((((....))))))))))))..)).((...(((((((....)))))))((((((.....)))-.)-------)).....))....---- ( -19.20) >DroPer_CAF1 75561 101 + 1 AACGUUUUAAUAAAUUUGUAAACAGAUUUGUUAAUUUCAGCAUAAAAUUUUGAUGAGAAUUUGGCCAUAUUGUGCA-UAAGACUCU-CUUGGAGACAGGAUCU ...(((((((((((((((....)))))))))........((((((((((((....))))))........)))))).-))))))..(-((((....)))))... ( -21.40) >consensus AACGUUUUAAUAAAUUUGUAAACAGAUUUGUUAAUUUCAGCAUAAAAUUUUGAUGAGAAUUUGGCCAUAUUGUGCA_UGA__CUCU_CAUGAAGACAGC____ ......((((((((((((....))))))))))))(((((.((........)).))))).....(((.....).))............................ (-12.66 = -12.30 + -0.36)
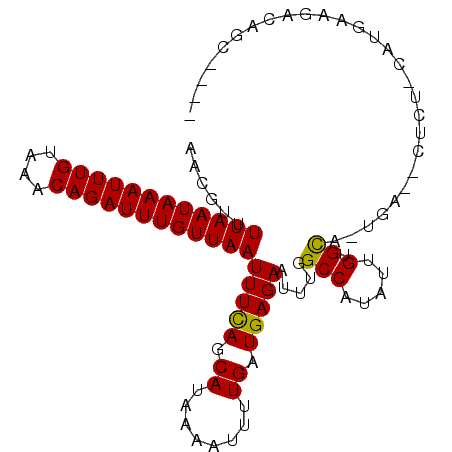
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:23 2006