
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,808,082 – 8,808,173 |
| Length | 91 |
| Max. P | 0.918618 |

| Location | 8,808,082 – 8,808,173 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -19.62 |
| Energy contribution | -19.90 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
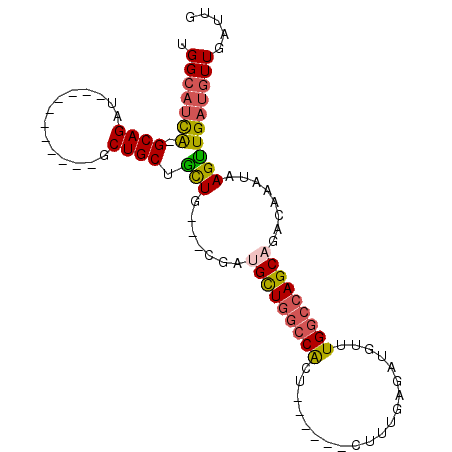
>2R_DroMel_CAF1 8808082 91 + 20766785 UGGCAUCA-GCAGAU-----------GCUGCUGCUGCUCCGAUGCUGGCCACUUUGAGACUUUGAGAUGUUUGGCCAGCAGACAAAUAAGUUGAUGUUGAUUG .(((((((-((...(-----------(..((....))..)).(((((((((......(((........)))))))))))).........)))))))))..... ( -29.90) >DroSec_CAF1 52593 88 + 1 UGGCAUCA-GCAGAU-----------GCUGCUGCUG---CGAUGCUGGCCACUUUGAGACUUUGAGAUGUUUGGCCAGCAGACAAAUAAGUUGAUGUUGAUUG .(((((((-((....-----------..(((....)---)).(((((((((......(((........)))))))))))).........)))))))))..... ( -30.00) >DroSim_CAF1 53703 88 + 1 UGGCAUCA-GCAGAU-----------GCUGCUGCUG---CGAUGCUGGCCACUUUGAGACUUUGAGAUGUUUGGCCAGCAGACAAAUAAGUUGAUGUUGAUUG .(((((((-((....-----------..(((....)---)).(((((((((......(((........)))))))))))).........)))))))))..... ( -30.00) >DroEre_CAF1 55465 94 + 1 UGGCAUCA-GCAGAUCCUGCUGCUGUGCUGCUGCUGCUGCGACGUUGGCCACU--------UUGAGAUGUUUGGCCAGCAGACAAAUAAGUUGAUGUUGAUUG .((((.((-((((......))))))))))((....))..((((((((((((..--------(......)..)))))))).(((......)))...)))).... ( -31.80) >DroYak_CAF1 55423 83 + 1 UGGAUUUA-GCAGAU-----------GCUGCUAUUUCUGCGAUGCUGGCCACU--------UUGAGAUGUUUGGCCAGCAGACAAAUAAGUUGAUGUUGAUUG .(((..((-((((..-----------.))))))..))).((((((((((((..--------(......)..)))))))).(((......)))...)))).... ( -26.30) >DroPer_CAF1 61195 74 + 1 UGGCAACGGGCAGAU-----------GCUGCUGCUG-------GUU---CG--------CUUUGAAAUGUUUGGCCAGCAGACAAAUAAGCUGAUGUUGAUUG ...(((((..(((..-----------..((((((((-------(((---.(--------(........))..))))))))).))......))).))))).... ( -22.80) >consensus UGGCAUCA_GCAGAU___________GCUGCUGCUG___CGAUGCUGGCCACU______CUUUGAGAUGUUUGGCCAGCAGACAAAUAAGUUGAUGUUGAUUG .(((((((.((((..............)))).(((.......(((((((((....................)))))))))........))))))))))..... (-19.62 = -19.90 + 0.28)

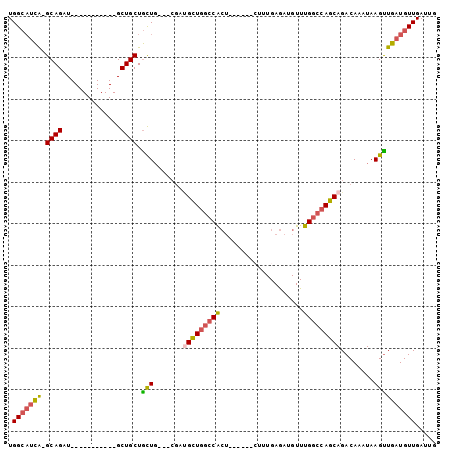
| Location | 8,808,082 – 8,808,173 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 80.59 |
| Mean single sequence MFE | -24.56 |
| Consensus MFE | -11.62 |
| Energy contribution | -13.51 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.47 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8808082 91 - 20766785 CAAUCAACAUCAACUUAUUUGUCUGCUGGCCAAACAUCUCAAAGUCUCAAAGUGGCCAGCAUCGGAGCAGCAGCAGC-----------AUCUGC-UGAUGCCA .................((((..(((((((((....................))))))))).))))(((.((((((.-----------..))))-)).))).. ( -27.65) >DroSec_CAF1 52593 88 - 1 CAAUCAACAUCAACUUAUUUGUCUGCUGGCCAAACAUCUCAAAGUCUCAAAGUGGCCAGCAUCG---CAGCAGCAGC-----------AUCUGC-UGAUGCCA .......................(((((((((....................)))))))))..(---((.((((((.-----------..))))-)).))).. ( -26.65) >DroSim_CAF1 53703 88 - 1 CAAUCAACAUCAACUUAUUUGUCUGCUGGCCAAACAUCUCAAAGUCUCAAAGUGGCCAGCAUCG---CAGCAGCAGC-----------AUCUGC-UGAUGCCA .......................(((((((((....................)))))))))..(---((.((((((.-----------..))))-)).))).. ( -26.65) >DroEre_CAF1 55465 94 - 1 CAAUCAACAUCAACUUAUUUGUCUGCUGGCCAAACAUCUCAA--------AGUGGCCAACGUCGCAGCAGCAGCAGCACAGCAGCAGGAUCUGC-UGAUGCCA .......(((((.......(((((((((((((..........--------..)))))......((....))))))).)))((((......))))-)))))... ( -26.20) >DroYak_CAF1 55423 83 - 1 CAAUCAACAUCAACUUAUUUGUCUGCUGGCCAAACAUCUCAA--------AGUGGCCAGCAUCGCAGAAAUAGCAGC-----------AUCUGC-UAAAUCCA .................(((((.(((((((((..........--------..)))))))))..)))))..((((((.-----------..))))-))...... ( -25.00) >DroPer_CAF1 61195 74 - 1 CAAUCAACAUCAGCUUAUUUGUCUGCUGGCCAAACAUUUCAAAG--------CG---AAC-------CAGCAGCAGC-----------AUCUGCCCGUUGCCA ....((((....((..(((((.(((((((........(((....--------.)---)))-------))))))))).-----------))..))..))))... ( -15.20) >consensus CAAUCAACAUCAACUUAUUUGUCUGCUGGCCAAACAUCUCAAAG______AGUGGCCAGCAUCG___CAGCAGCAGC___________AUCUGC_UGAUGCCA .......(((((...........(((((((((....................)))))))))...........((((..............)))).)))))... (-11.62 = -13.51 + 1.89)


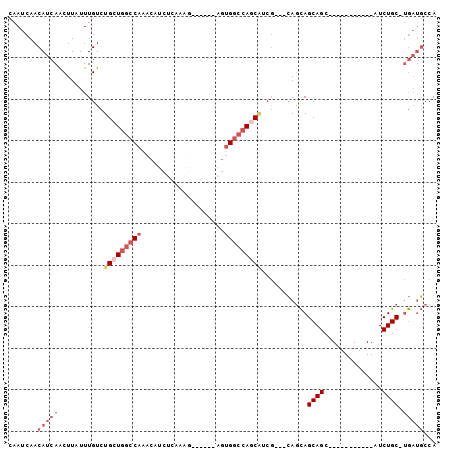
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:17 2006