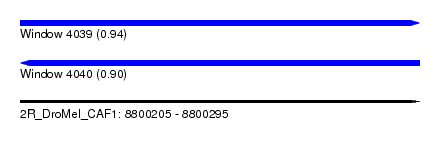
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,800,205 – 8,800,295 |
| Length | 90 |
| Max. P | 0.938578 |

| Location | 8,800,205 – 8,800,295 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 69.32 |
| Mean single sequence MFE | -18.67 |
| Consensus MFE | -10.71 |
| Energy contribution | -11.27 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938578 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
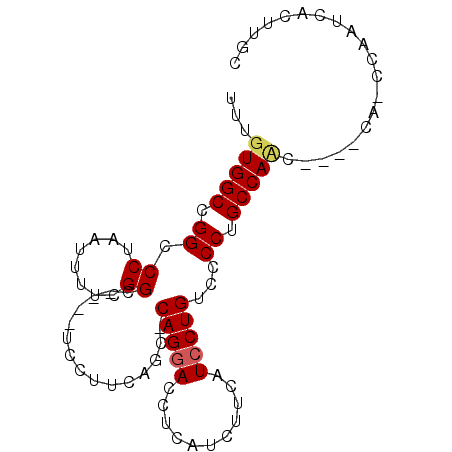
>2R_DroMel_CAF1 8800205 90 + 20766785 UUUGUGGCCGGCCCUAAUUUUCGGCAUUUUCCUUCAGC-CAGGAUCUCAGCUUCAUCCUGUGCCCUGCCAUCCUGCCAUCCAAUCACUUGC ...((((((((.........)))))...........(.-((((((..(((...(((...)))..)))..)))))).).......))).... ( -18.20) >DroSim_CAF1 45863 75 + 1 UUUGUGGCUGGCCCUUAUUU-CGG----------CAGC-CAGGACCUCAUCUUCUUGCUGUUCCCUGCCACC----CACCCAAUCACUUGC ...((((.((((........-.((----------((((-.((((.......)))).))))))....)))).)----)))............ ( -18.82) >DroEre_CAF1 47867 80 + 1 UUUGUGGCCGGCCCAAAUGCUCGGC----UCCUUCAGCCCAGGACCUCAUCUUCAUCCUGGCCCCUGCCAAU-------UUUUGCUUUUGC ....((((.((((.........(((----(.....)))).((((...........))))))))...))))..-------............ ( -19.00) >consensus UUUGUGGCCGGCCCUAAUUUUCGGC____UCCUUCAGC_CAGGACCUCAUCUUCAUCCUGUCCCCUGCCAAC____CA_CCAAUCACUUGC ...(((((.((.((........))...............(((((...........)))))...)).))))).................... (-10.71 = -11.27 + 0.56)


| Location | 8,800,205 – 8,800,295 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 69.32 |
| Mean single sequence MFE | -24.47 |
| Consensus MFE | -12.64 |
| Energy contribution | -13.20 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
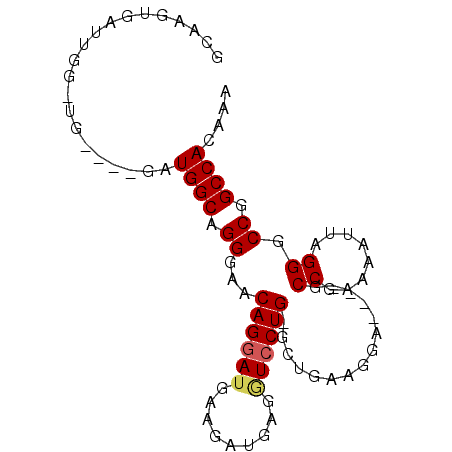
>2R_DroMel_CAF1 8800205 90 - 20766785 GCAAGUGAUUGGAUGGCAGGAUGGCAGGGCACAGGAUGAAGCUGAGAUCCUG-GCUGAAGGAAAAUGCCGAAAAUUAGGGCCGGCCACAAA .(((....)))..((((....((((..(((.((((((.........))))))-)))...........((........)))))))))).... ( -24.00) >DroSim_CAF1 45863 75 - 1 GCAAGUGAUUGGGUG----GGUGGCAGGGAACAGCAAGAAGAUGAGGUCCUG-GCUG----------CCG-AAAUAAGGGCCAGCCACAAA .(((....))).(((----(.((((......((((....((........)).-))))----------((.-......)))))).))))... ( -23.50) >DroEre_CAF1 47867 80 - 1 GCAAAAGCAAAA-------AUUGGCAGGGGCCAGGAUGAAGAUGAGGUCCUGGGCUGAAGGA----GCCGAGCAUUUGGGCCGGCCACAAA ((....))....-------..((((...((((..((((..(((...)))((.((((.....)----))).))))))..)))).)))).... ( -25.90) >consensus GCAAGUGAUUGG_UG____GAUGGCAGGGAACAGGAUGAAGAUGAGGUCCUG_GCUGAAGGA____GCCGAAAAUUAGGGCCGGCCACAAA .....................((((.((...((((((.........))))))...............((........)).)).)))).... (-12.64 = -13.20 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:14 2006