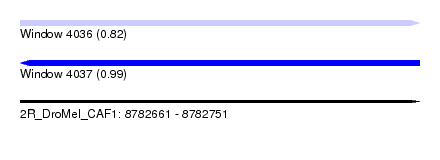
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,782,661 – 8,782,751 |
| Length | 90 |
| Max. P | 0.989484 |

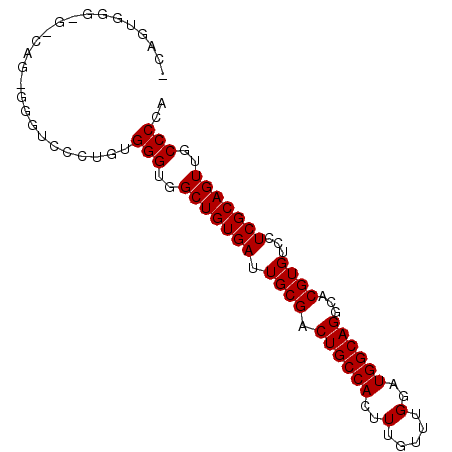
| Location | 8,782,661 – 8,782,751 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 86.43 |
| Mean single sequence MFE | -28.52 |
| Consensus MFE | -22.68 |
| Energy contribution | -22.68 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8782661 90 + 20766785 UGGGGCAACUGCGAGGACACGUGCCUGCCAUCCAAACAAAGUGGCAGUCGCAAUCACAGCCACCCACAGGGACCCUCUGUCUCCCACUGC ((((((....))((((....(((.(((((((.........))))))).)))...........(((...)))..)))).....)))).... ( -29.10) >DroSec_CAF1 27030 90 + 1 UGGGGCAACUGCGAGGACACGUGCCUGCCAUCCAAACAAAGUGGCAGUCGCAAUCACAGCCACCCACAGGGACUCGCUGCCGCCCACUGU ..((((....(((((.....(((.(((((((.........))))))).)))...........(((...))).)))))....))))..... ( -33.80) >DroSim_CAF1 28179 90 + 1 UGGGGCAACUGCGAGGACACGUGCCUGCCAUCCAAACAAAGUGGCAGUCGCAAUCACAGCCACCCACAGGGACUCGCUGCCGCCCACUGU ..((((....(((((.....(((.(((((((.........))))))).)))...........(((...))).)))))....))))..... ( -33.80) >DroEre_CAF1 28687 74 + 1 UGGGGCAACUGCGAGGACACGUGCCUGCCAUACAAACAAAGUGGCAGUCGCAAUCACAGCCACCCACAGGGACC---------------- ((((....(((.(((....)(((.(((((((.........))))))).)))..)).)))...))))........---------------- ( -22.80) >DroYak_CAF1 29927 74 + 1 UGGGGCAACUGCGAGGACACGUGCCUGCCAUACAAACAAAGUGGCAGUCGCAAUCACAGCCACCCAAAGGGACC---------------- ((((....(((.(((....)(((.(((((((.........))))))).)))..)).)))...))))........---------------- ( -23.10) >consensus UGGGGCAACUGCGAGGACACGUGCCUGCCAUCCAAACAAAGUGGCAGUCGCAAUCACAGCCACCCACAGGGACCC_CUG_C_CCCACUG_ ((((....(((.(((....)(((.(((((((.........))))))).)))..)).)))...))))........................ (-22.68 = -22.68 + -0.00)

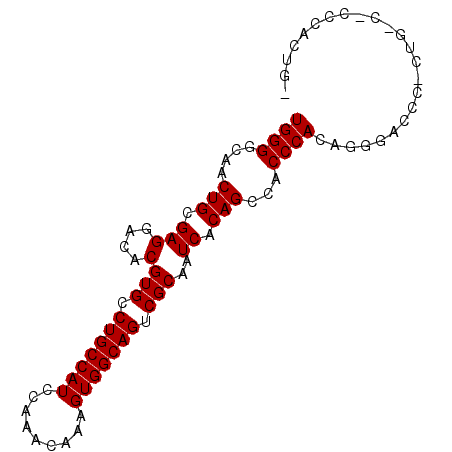
| Location | 8,782,661 – 8,782,751 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 86.43 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -30.88 |
| Energy contribution | -30.88 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8782661 90 - 20766785 GCAGUGGGAGACAGAGGGUCCCUGUGGGUGGCUGUGAUUGCGACUGCCACUUUGUUUGGAUGGCAGGCACGUGUCCUCGCAGUUGCCCCA ...(..((.(((.....)))))..)(((..(((((((.((((.((((((((......)).))))))...))))...)))))))..))).. ( -39.60) >DroSec_CAF1 27030 90 - 1 ACAGUGGGCGGCAGCGAGUCCCUGUGGGUGGCUGUGAUUGCGACUGCCACUUUGUUUGGAUGGCAGGCACGUGUCCUCGCAGUUGCCCCA ((((.((((........))))))))(((..(((((((.((((.((((((((......)).))))))...))))...)))))))..))).. ( -42.80) >DroSim_CAF1 28179 90 - 1 ACAGUGGGCGGCAGCGAGUCCCUGUGGGUGGCUGUGAUUGCGACUGCCACUUUGUUUGGAUGGCAGGCACGUGUCCUCGCAGUUGCCCCA ((((.((((........))))))))(((..(((((((.((((.((((((((......)).))))))...))))...)))))))..))).. ( -42.80) >DroEre_CAF1 28687 74 - 1 ----------------GGUCCCUGUGGGUGGCUGUGAUUGCGACUGCCACUUUGUUUGUAUGGCAGGCACGUGUCCUCGCAGUUGCCCCA ----------------.......(.(((..(((((((.((((.((((((..(.....)..))))))...))))...)))))))..)))). ( -31.10) >DroYak_CAF1 29927 74 - 1 ----------------GGUCCCUUUGGGUGGCUGUGAUUGCGACUGCCACUUUGUUUGUAUGGCAGGCACGUGUCCUCGCAGUUGCCCCA ----------------.........(((..(((((((.((((.((((((..(.....)..))))))...))))...)))))))..))).. ( -30.70) >consensus _CAGUGGG_G_CAG_GGGUCCCUGUGGGUGGCUGUGAUUGCGACUGCCACUUUGUUUGGAUGGCAGGCACGUGUCCUCGCAGUUGCCCCA .........................(((..(((((((.((((.((((((..(.....)..))))))...))))...)))))))..))).. (-30.88 = -30.88 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:11 2006