| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,773,617 – 8,773,722 |
| Length | 105 |
| Max. P | 0.532531 |

| Location | 8,773,617 – 8,773,722 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.39 |
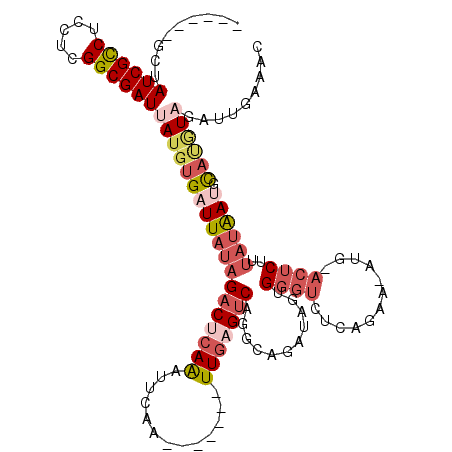
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -13.95 |
| Energy contribution | -15.87 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.52 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
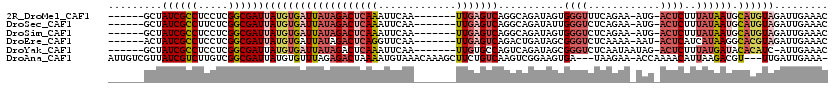
>2R_DroMel_CAF1 8773617 105 - 20766785 ------GCUAUCGCCUCCUCGGCGAUUAUGUGAUUAUAGACUCAAAUUCAA-------UUGAGUCAGGCAGAUAGUGGGUUUCAGAA-AUG-ACUCUUUAUAAUGCAUGUAGAUUGAAAC ------....(((((.....)))))((((((((((((((((((((......-------)))))))...........(((..(((...-.))-)..))))))))).)))))))........ ( -27.50) >DroSec_CAF1 18112 105 - 1 ------GCUAUCGCCUUCUCGGCGAUUAUGUGAUUAUAGACUCAAAUUCAA-------UUGAGUCAGGCAGAUAUUGGGUCUCAGAA-AUG-ACUCUUUAUAAUGCAUGUAGAUUGAAAC ------((.((((((.....))))))...))..((((((((((((......-------)))))))..(((.(((..(((((......-..)-))))..)))..))).)))))........ ( -29.80) >DroSim_CAF1 18700 105 - 1 ------GCUAUCGCCUCCUCGGCGAUUAUGUGAUUAUAGACUCAAAUUCAA-------UUGAGUCAGGCAGAUAGUGGGUCUCAGAA-AUG-ACUCUUUAUAAUGCAUGUAGAUUGAAAC ------....(((((.....)))))((((((((((((((((((((......-------)))))))...........(((((......-..)-))))..)))))).)))))))........ ( -29.70) >DroEre_CAF1 20370 105 - 1 ------ACUAUCGCCUCCUCGGCGAUUAUGUGAUUAUAGACUCAGGUUCAA-------UUGAGUCAGACUGAUAGCGGGUCUCAAAA-AAU-ACUCAUCAUAAGGCACGUAGAUUGAAAC ------....(((((.....)))))(((((((.((((.(((((((......-------)))))))(((((.......))))).....-...-.......))))..)))))))........ ( -27.60) >DroYak_CAF1 20569 105 - 1 ------GCUAUCGCCUCCUCGGCGAUUAUGUGAUUAUAGACUCAAAUUCAA-------UUGUGCCAGUCAGAUAGCGGGUCUCAAUAAUAG-ACUCUUUAUGAUACACAUC-AUUGAAAC ------...((((((.....)))))).(((((((((((((((...((....-------..))...)))).......((((((.......))-))))..)))))).))))).-........ ( -24.80) >DroAna_CAF1 19934 112 - 1 AUUGUCGUUAUCGUCUUGUCGGCGAUUAUGUGUUUAGAGACUAAAAUGUAAACAAAGCUUCUGUCAAGUCGGAAGUGA---UAAGAA-ACCAAAACAUUAAGACGU---UUGAUUGAAA- ...((((....(((((((..(.........((((((.(........).))))))..(((((((......)))))))..---......-.......)..))))))).---.)))).....- ( -21.40) >consensus ______GCUAUCGCCUCCUCGGCGAUUAUGUGAUUAUAGACUCAAAUUCAA_______UUGAGUCAGGCAGAUAGUGGGUCUCAGAA_AUG_ACUCUUUAUAAUGCAUGUAGAUUGAAAC .........((((((.....))))))(((((((((((((((((((.............)))))))...........((((............))))..)))))).))))))......... (-13.95 = -15.87 + 1.92)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:07 2006