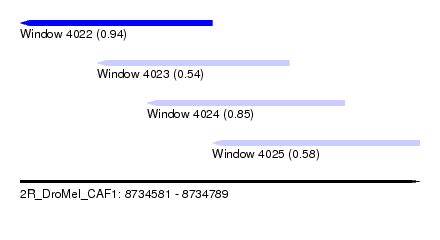
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,734,581 – 8,734,789 |
| Length | 208 |
| Max. P | 0.940506 |

| Location | 8,734,581 – 8,734,681 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 89.00 |
| Mean single sequence MFE | -23.90 |
| Consensus MFE | -22.20 |
| Energy contribution | -21.45 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8734581 100 - 20766785 UUACGGCACUUGGAAGACGUAUUC------CCAUUACAACACUCUACGCUUUCCAAGAUGCAGCAUUCUAUCUCAAUUGACACUGAUUGGAUUGCGUGCAUGCAUA ........(((((((..((((...------..............))))..)))))))(((((((((.(.(((.(((((......)))))))).).)))).))))). ( -28.03) >DroSec_CAF1 3693 100 - 1 CUACGGCUCUUGGAAGACAUAUUC------CCAUUACAACACUCUACGCUUUCCAAGAUGCAGCAUUCUAUCUCAAUUGACAAUGAUUGGAUUGCGUGCAUGCAUA .......((((((((..(......------.................)..))))))))((((((((.(.(((.(((((......)))))))).).)))).)))).. ( -22.70) >DroSim_CAF1 7196 100 - 1 CUACGGCACUUGGAAGACAUAUUC------CCAUUACAACACUCUACGCUUUCCAAGAUGCAGCAUUCUAUCUCAAUUGACAAUGAUUGGAUUGCGUGCAUGCAUA ........(((((((..(......------.................)..)))))))(((((((((.(.(((.(((((......)))))))).).)))).))))). ( -23.00) >DroYak_CAF1 4598 104 - 1 CUGGAACACUUGGAAGACUUAUUCCCAUUCCCAUUACAACACU--UCGCUUUCUGGGAUGCAGCAUUUUAUCUCAAUUGACAAUGAUUGGAUUGCGUGCCUGCAUA ...........((((......))))...(((((..........--........)))))((((((((...(((.(((((......))))))))...))).))))).. ( -21.87) >consensus CUACGGCACUUGGAAGACAUAUUC______CCAUUACAACACUCUACGCUUUCCAAGAUGCAGCAUUCUAUCUCAAUUGACAAUGAUUGGAUUGCGUGCAUGCAUA ........(((((((..(.............................)..)))))))(((((((((.(.(((.(((((......)))))))).).)))).))))). (-22.20 = -21.45 + -0.75)

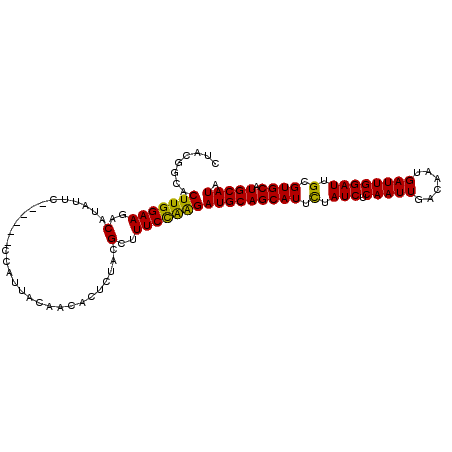
| Location | 8,734,621 – 8,734,721 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 87.22 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -17.39 |
| Energy contribution | -17.20 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8734621 100 - 20766785 GGCUGGCACGCAUCAGUCAUGCAUCUUCUCCAUUUGUCAAUUACGGCACUUGGAAGACGUAUUC------CCAUUACAACACUCUACGCUUUCCAAGAUGCAGCAU ((((((......)))))).((((((((.((((..((((......))))..))))((.((((...------..............))))))....)))))))).... ( -25.83) >DroSec_CAF1 3733 100 - 1 AGCUGGCACGCAUCAAUCAUGCAUCUUCUCCGUUUGUCAACUACGGCUCUUGGAAGACAUAUUC------CCAUUACAACACUCUACGCUUUCCAAGAUGCAGCAU .((((.((.((((.....)))).......((((.........)))).((((((((..(......------.................)..)))))))))))))).. ( -20.40) >DroSim_CAF1 7236 100 - 1 GGCUGGCACGCAUCAGUCAUGCAUCUUCUCCGUUUGUCAACUACGGCACUUGGAAGACAUAUUC------CCAUUACAACACUCUACGCUUUCCAAGAUGCAGCAU ((((((......)))))).((((((((.((((..((((......))))..))))(((.......------............))).........)))))))).... ( -22.41) >DroYak_CAF1 4638 104 - 1 GGCUGGCACGCUUCAAUCAUGCAUCUUCUCCAGUUGUUAACUGGAACACUUGGAAGACUUAUUCCCAUUCCCAUUACAACACU--UCGCUUUCUGGGAUGCAGCAU .((((.((.((.........))......((((((.....))))))......((((......))))...(((((..........--........))))))))))).. ( -21.77) >consensus GGCUGGCACGCAUCAAUCAUGCAUCUUCUCCAUUUGUCAACUACGGCACUUGGAAGACAUAUUC______CCAUUACAACACUCUACGCUUUCCAAGAUGCAGCAU ((((((......)))))).((((((((.((((..((((......))))..)))).((.....))..............................)))))))).... (-17.39 = -17.20 + -0.19)

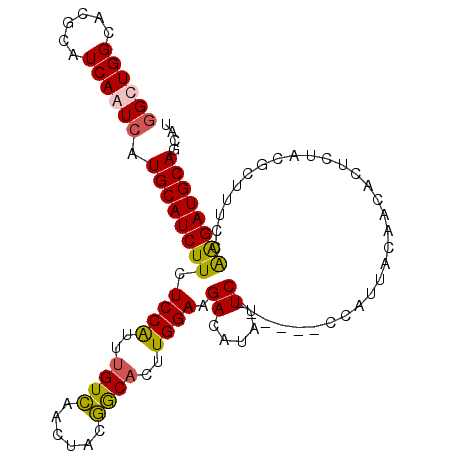

| Location | 8,734,647 – 8,734,750 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 72.06 |
| Mean single sequence MFE | -24.01 |
| Consensus MFE | -6.42 |
| Energy contribution | -8.14 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.27 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.847976 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8734647 103 - 20766785 ACACGGUUCAUCCAUAAUGCAU----GAUUCUAGGCUGGCACGCAUCAGUCAUGCAUCUUCUCCAUUUGUCAAUUACGGCACUUGGAAGACGUAUUC------CCAUUACAAC ..(((...........((((((----((((....((......))...))))))))))..((((((..((((......))))..))).))))))....------.......... ( -25.10) >DroSec_CAF1 3759 103 - 1 ACACGGUUCAUCCAUAAUGCAU----GAUUCUAAGCUGGCACGCAUCAAUCAUGCAUCUUCUCCGUUUGUCAACUACGGCUCUUGGAAGACAUAUUC------CCAUUACAAC ....((...........(((((----((((....((......))...)))))))))((((((((((.........)))).....)))))).......------))........ ( -21.20) >DroSim_CAF1 7262 103 - 1 ACACGGUUCAUCCAUAAUGCAU----GAUUCUAGGCUGGCACGCAUCAGUCAUGCAUCUUCUCCGUUUGUCAACUACGGCACUUGGAAGACAUAUUC------CCAUUACAAC ....((..........((((((----((((....((......))...))))))))))..((((((..((((......))))..))).))).......------))........ ( -23.70) >DroYak_CAF1 4662 109 - 1 ACACGGUUCAUCCAUUAUGCAU----GAUUCUAGGCUGGCACGCUUCAAUCAUGCAUCUUCUCCAGUUGUUAACUGGAACACUUGGAAGACUUAUUCCCAUUCCCAUUACAAC ....((((..((((..((((((----((((..((((......)))).))))))))))....((((((.....)))))).....)))).))))..................... ( -27.10) >DroPer_CAF1 3282 87 - 1 CCAUGGCUCAUCCAUAAUGCAGUCAUAUUGCAUAGUCAGCACGCAUCAAUCAUCUAGAGCUGGCCUGCGGCAGCUACUGC-----------------------CCGAU---GC ..(((((((((.....))).))))))...(((..((((((.(..............).)))))).)))(((((...))))-----------------------)....---.. ( -22.94) >consensus ACACGGUUCAUCCAUAAUGCAU____GAUUCUAGGCUGGCACGCAUCAAUCAUGCAUCUUCUCCAUUUGUCAACUACGGCACUUGGAAGACAUAUUC______CCAUUACAAC ....((..........((((((....((((....((......))...))))))))))....((((..((((......))))..))))................))........ ( -6.42 = -8.14 + 1.72)

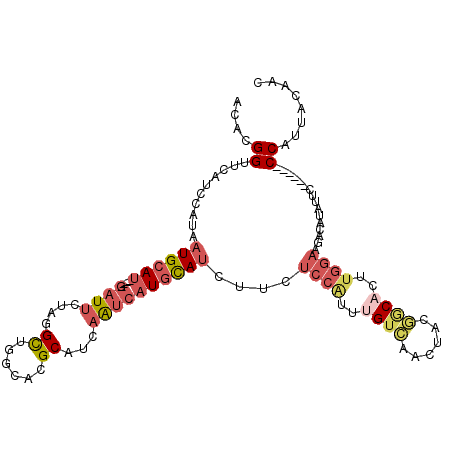
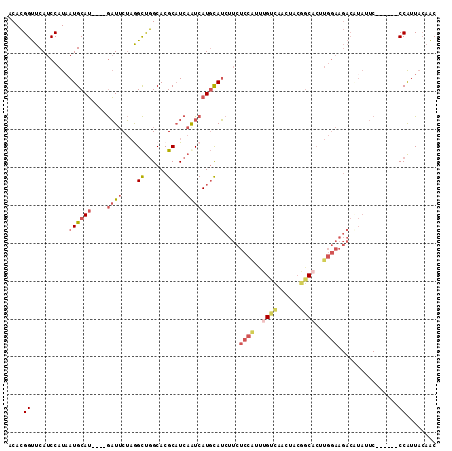
| Location | 8,734,681 – 8,734,789 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 94.14 |
| Mean single sequence MFE | -24.26 |
| Consensus MFE | -19.66 |
| Energy contribution | -20.23 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8734681 108 - 20766785 GAAUACUCAUAUCAAGCGGUCUACCCCUGGCCACUUCCCACACGGUUCAUCCAUAAUGCAUGAUUCUAGGCUGGCACGCAUCAGUCAUGCAUCUUCUCCAUUUGUCAA ((((.........(((.((((.......)))).)))........)))).......((((((((((....((......))...))))))))))................ ( -22.63) >DroSec_CAF1 3793 108 - 1 GAAUACUCAUAUCAAGCGGUCUACCCCUGGACACUUCCCACACGGUUCAUCCAUAAUGCAUGAUUCUAAGCUGGCACGCAUCAAUCAUGCAUCUUCUCCGUUUGUCAA ............(((((((........((((.....((.....))....))))..((((((((((....((......))...)))))))))).....))))))).... ( -24.50) >DroSim_CAF1 7296 108 - 1 GAAUACUCAUAUCAAGCGGUCUACCCCUGGCCACUUCCCACACGGUUCAUCCAUAAUGCAUGAUUCUAGGCUGGCACGCAUCAGUCAUGCAUCUUCUCCGUUUGUCAA ............(((((((...(((..(((.......)))...))).........((((((((((....((......))...)))))))))).....))))))).... ( -24.40) >DroYak_CAF1 4702 108 - 1 GAAUGCUCAUAUCGAGCGCUUUACCCCUGGCCACUUCCCACACGGUUCAUCCAUUAUGCAUGAUUCUAGGCUGGCACGCUUCAAUCAUGCAUCUUCUCCAGUUGUUAA ....((((.....)))).........((((.............((.....))...((((((((((..((((......)))).)))))))))).....))))....... ( -25.50) >consensus GAAUACUCAUAUCAAGCGGUCUACCCCUGGCCACUUCCCACACGGUUCAUCCAUAAUGCAUGAUUCUAGGCUGGCACGCAUCAAUCAUGCAUCUUCUCCAUUUGUCAA ............(((((((...(((..(((.......)))...))).........((((((((((....((......))...)))))))))).....))))))).... (-19.66 = -20.23 + 0.56)

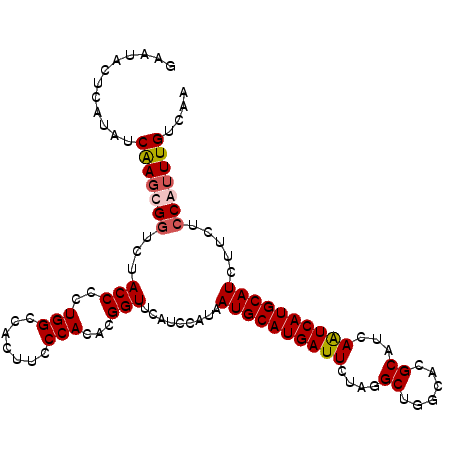
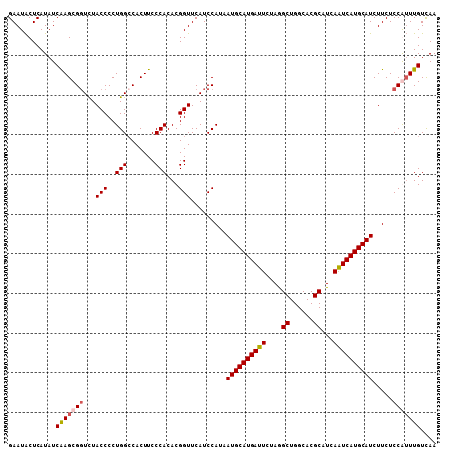
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:30:00 2006