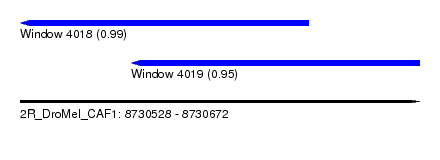
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,730,528 – 8,730,672 |
| Length | 144 |
| Max. P | 0.990771 |

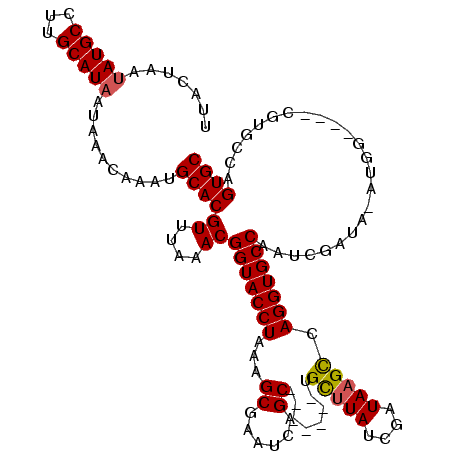
| Location | 8,730,528 – 8,730,632 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.64 |
| Mean single sequence MFE | -36.27 |
| Consensus MFE | -19.37 |
| Energy contribution | -19.27 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8730528 104 - 20766785 ACGGUACCUAAAGCGAAUCAGC-------UGCUUAUCGAUAAGUCAGGUGCCAAUCGAUACAUGG---------CAGUGCUGCCCAGUAUCGCACAGCAAGGUGCGUUUAAAUUUUGAAA ..(((((((..(((......))-------)(((((....))))).)))))))..(((((((..((---------((....))))..)))))((((......))))...........)).. ( -33.70) >DroSim_CAF1 537 111 - 1 ACGGUACCUAAAGCGAAUCAGC-------UGCUUAACGAUAAGCCAGGUGCCAAUCGAUAUAUGGGCCACGUGCCAGUGCUGCCCA-UAUCGCACAGCAAAGUGCGUAUA-AUUUUGAAA ..(((((((..(((......))-------)(((((....))))).)))))))..((((.(((((((((((......)))..)))))-)))(((((......)))))....-...)))).. ( -40.30) >DroYak_CAF1 559 105 - 1 ACGGUACCUAAAGCCAAUCAGCUGGCAGCUGUCUAUCGAUAAGCCAGGUGCCGC---------------CGUGCCAGUGCUGCCCAGCGUAGCUCAGCAAACUGCGGUUACUUUUUGAAA .((((((((...((((......)))).((((((....))).))).)))))))).---------------.(((((.(.(((((.....))))).).((.....)))).)))......... ( -34.80) >consensus ACGGUACCUAAAGCGAAUCAGC_______UGCUUAUCGAUAAGCCAGGUGCCAAUCGAUA_AUGG____CGUGCCAGUGCUGCCCAGUAUCGCACAGCAAAGUGCGUUUA_AUUUUGAAA ..(((((((...((......))........(((((....))))).))))))).........................(((((..(......)..)))))..................... (-19.37 = -19.27 + -0.11)

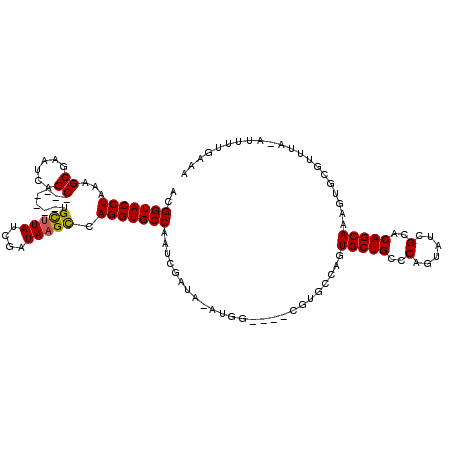
| Location | 8,730,568 – 8,730,672 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.36 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -20.11 |
| Energy contribution | -20.33 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8730568 104 - 20766785 UUACAAAUAUGCCUUGCAUAAUAAACAAAUGCACGUUUAAACGGUACCUAAAGCGAAUCAGC-------UGCUUAUCGAUAAGUCAGGUGCCAAUCGAUACAUGG---------CAGUGC .........((((.(((((.........))))).........(((((((..(((......))-------)(((((....))))).)))))))...........))---------)).... ( -25.40) >DroSim_CAF1 575 113 - 1 UUACUAAUAUGCCUUGCAUAAUAAACAAAUGCACGUUUAAACGGUACCUAAAGCGAAUCAGC-------UGCUUAACGAUAAGCCAGGUGCCAAUCGAUAUAUGGGCCACGUGCCAGUGC .......(((((...)))))..........((((........(((((((..(((......))-------)(((((....))))).)))))))............(((.....))).)))) ( -29.70) >DroYak_CAF1 599 105 - 1 UUACUAACAUGCCUUGCAUAAUAAACAAACGCACGUGUGAACGGUACCUAAAGCCAAUCAGCUGGCAGCUGUCUAUCGAUAAGCCAGGUGCCGC---------------CGUGCCAGUGC .((((...((((...))))...........(((((.(....((((((((...((((......)))).((((((....))).))).)))))))))---------------))))).)))). ( -31.70) >consensus UUACUAAUAUGCCUUGCAUAAUAAACAAAUGCACGUUUAAACGGUACCUAAAGCGAAUCAGC_______UGCUUAUCGAUAAGCCAGGUGCCAAUCGAUA_AUGG____CGUGCCAGUGC .......(((((...)))))..........((((((....))(((((((...((......))........(((((....))))).)))))))........................)))) (-20.11 = -20.33 + 0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:54 2006