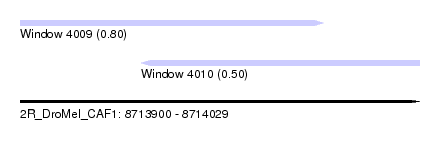
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,713,900 – 8,714,029 |
| Length | 129 |
| Max. P | 0.803981 |

| Location | 8,713,900 – 8,713,998 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.33 |
| Mean single sequence MFE | -30.76 |
| Consensus MFE | -12.28 |
| Energy contribution | -12.88 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.40 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
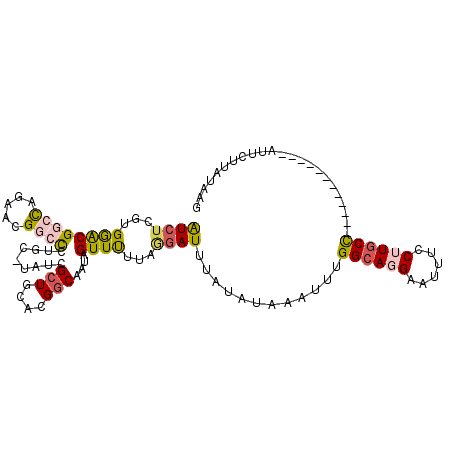
>2R_DroMel_CAF1 8713900 98 + 20766785 AUCUCGUGAGCGGCUAGAACGGCCAUGC-UAUCGCUGCACGACAGUAGUUCUUAGAAUUUAUAUAAAUUUGGCAGGAAUUUCCUUGCC------------AUUCUUAUAGG .(((...((((((((.....)))).(((-(.(((.....))).))))))))..))).....(((((...(((((((......))))))------------)...))))).. ( -28.10) >DroSec_CAF1 20122 108 + 1 GUCUCGUGGACGACCAGAACGGCCCUGC-UAUCGCUGCACGGCAAUAGUUCUUAAGAUUUAUAUAAAUUUGGCAGGAAU--CCGUUCUUAUAAGAAGAUGAUUCUUAUAAG (((.....))).....((((((.(((((-((..(((....))).((((.((....)).)))).......)))))))...--))))))(((((((((.....))))))))). ( -32.30) >DroSim_CAF1 22976 98 + 1 GUCUCGUGGACGGCCAGAACGGCCCUGC-UAUCGCUGCACGGCAAUAGUUUUUAGGAUUUACAUAAAUUUGGCAGGAAUUUCCGUGCC------------AUUCUUAUAAA .....(((((.((((.....))))..((-(((.(((....))).)))))........)))))((((...(((((((.....)).))))------------)...))))... ( -26.60) >DroEre_CAF1 20700 97 + 1 AUCUCGUGGACGGCCAGAACGCCCUGG--CCUUGCUGCACGGCAAUAGUUUUUACGAUUUAUAUAUAUUUGGCAGGGAUUUCCUUGCC------------AUUCUUAUAAG ...(((((((.((((((......))))--))(((((....))))).....)))))))....((((....((((((((....)))))))------------)....)))).. ( -37.10) >DroYak_CAF1 20773 99 + 1 AUCUCGUGGACGGCCAGAAGGCAUGUGCCCCUUGCUGCACGGCAUUAGUUUUUAGGAUUUACAUAUAUUUGGCAGGGAUUUCCUUGCC------------AUUCUUAUAAG .....(((((...((.((((((..(((((...........)))))..)))))).)).))))).((((..((((((((....)))))))------------)....)))).. ( -29.70) >consensus AUCUCGUGGACGGCCAGAACGGCCCUGC_UAUCGCUGCACGGCAAUAGUUUUUAGGAUUUAUAUAAAUUUGGCAGGAAUUUCCUUGCC____________AUUCUUAUAAG ((((...((((((((.....)))).........(((....)))....))))...))))............((((((......))))))....................... (-12.28 = -12.88 + 0.60)

| Location | 8,713,939 – 8,714,029 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 85.86 |
| Mean single sequence MFE | -19.92 |
| Consensus MFE | -13.78 |
| Energy contribution | -13.38 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
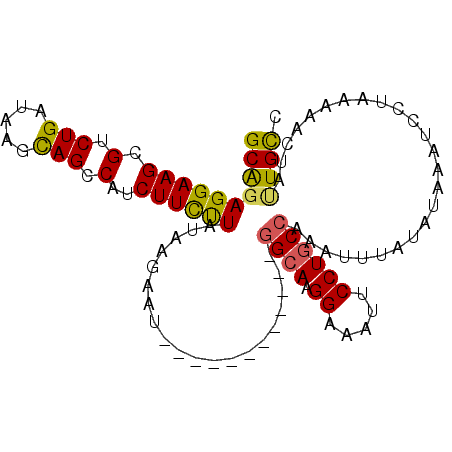
>2R_DroMel_CAF1 8713939 90 - 20766785 GCAGAGGAAGCGUCUGAUAAGCAGCCAUCUUCCUAUAAGAAU------------GGCAAGGAAAUUCCUGCCAAAUUUAUAUAAAUUCUAAGAACUACUGUC ((((((((((.(.(((.....))).)..))))))(((((..(------------((((.((.....)))))))..))))).................)))). ( -22.50) >DroSec_CAF1 20161 100 - 1 GCAGAGGAAGCGUCUGAUAAGUAGCCAUCUUCUUAUAAGAAUCAUCUUCUUAUAAGAACGG--AUUCCUGCCAAAUUUAUAUAAAUCUUAAGAACUAUUGCC .(((((....).))))....((((..(((((((((((((((.....)))))))))))..))--))..))))............................... ( -20.10) >DroSim_CAF1 23015 90 - 1 GCAGAGGAAGCGUCUGAUAAGUAGCCAUCUUUUUAUAAGAAU------------GGCACGGAAAUUCCUGCCAAAUUUAUGUAAAUCCUAAAAACUAUUGCC ((((((((.((..((....))..))......((((((.((((------------((((.((.....)))))))..))).))))))))))........)))). ( -17.90) >DroEre_CAF1 20738 90 - 1 GCGGAGGAAGCGUCUGAUAAGCAGCCAUCUUCUUAUAAGAAU------------GGCAAGGAAAUCCCUGCCAAAUAUAUAUAAAUCGUAAAAACUAUUGCC ((((((((.(.(.(((.....))))).))))))........(------------((((.((....)).)))))..........................)). ( -18.90) >DroYak_CAF1 20813 90 - 1 GCAGAGGAAGCGUCUGAUAAGCAGCCAUCUUCUUAUAAGAAU------------GGCAAGGAAAUCCCUGCCAAAUAUAUGUAAAUCCUAAAAACUAAUGCC (((.((((....(((.(((((.((....)).))))).))).(------------((((.((....)).)))))............)))).........))). ( -20.20) >consensus GCAGAGGAAGCGUCUGAUAAGCAGCCAUCUUCUUAUAAGAAU____________GGCAAGGAAAUUCCUGCCAAAUUUAUAUAAAUCCUAAAAACUAUUGCC ((((((((((.(.(((.....))).)..))))))....................((((.((.....)))))).........................)))). (-13.78 = -13.38 + -0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:46 2006