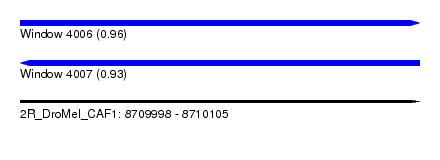
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,709,998 – 8,710,105 |
| Length | 107 |
| Max. P | 0.963285 |

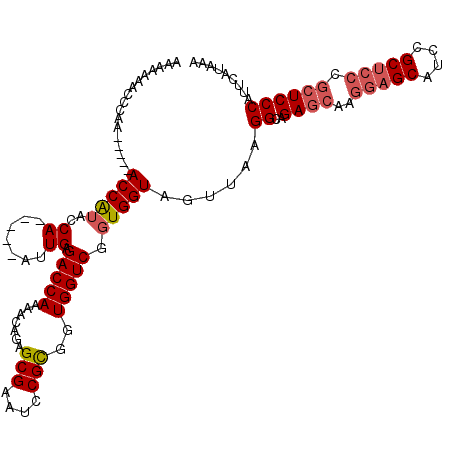
| Location | 8,709,998 – 8,710,105 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 90.76 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -28.96 |
| Energy contribution | -30.12 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8709998 107 + 20766785 UUUAUGAAUGGGAGCGGGAGCGGAUGCUCCUUGCUCUGCCUCAACUACCACCGACCACCGCGGAUUCGCUCUGUUUUGGUCUCAAU-----UGGUAUGGU-----UUGGGUUUUUUU ..........(((((((((((....))))).))))))((((.((((((((..(((((..(((((.....)))))..))))).....-----))))..)))-----).))))...... ( -38.20) >DroSec_CAF1 16178 106 + 1 UUUAUGAAUGGGAGCGGGAGCGGAUGCUCCUUGCUCUGCCUUAACUACCACCGACCACCGCGGAUUCGCUCUGUUUUGGUCUCAAU-----UGGUAUGGU-----UUGGGUUUU-UU ..........(((((((((((....))))).))))))((((.((((((((..(((((..(((((.....)))))..))))).....-----))))..)))-----).))))...-.. ( -38.20) >DroSim_CAF1 19159 107 + 1 UUUAUGAAUGGGAGCGGGAGCGGAUGCUCCUUGCUCUGCCUUAACUACCACCGACCACCGCGGAUUCGCUCUGUUUUGGUCUCAAU-----UGGUAUGGU-----UUGGGUUUUUUU ..........(((((((((((....))))).))))))((((.((((((((..(((((..(((((.....)))))..))))).....-----))))..)))-----).))))...... ( -38.20) >DroEre_CAF1 16955 106 + 1 UUUAUGAAUGGGA------GCGGAUGCUCCUUGCUCUGCCUUAACUACCACCGACCACCACGGAUUCGCUCUGUUUUGGUCUCAAU-----UGGUUCGGUUUGGUUUGGGGUUUUUU .........((((------((....)))))).((((((((..((((((((..(((((..(((((.....)))))..))))).....-----))))..)))).)))..)))))..... ( -31.50) >DroYak_CAF1 16946 117 + 1 UUUAUGAAUGGGAGCGGGAGCGGAUGCUCCUUGCUCUGCCUUAACUACCACCGACCACCACGGAUUCGCUCUGUUUUGGUCUCAAUAUGUAUGGUAUGGUUUGGUUUGGGUUCUUUU ..........(((((((((((....))))).))))))((((.(((((..((((((((..(((((.....)))))..)))))...((((.....))))))).))))).))))...... ( -38.90) >consensus UUUAUGAAUGGGAGCGGGAGCGGAUGCUCCUUGCUCUGCCUUAACUACCACCGACCACCGCGGAUUCGCUCUGUUUUGGUCUCAAU_____UGGUAUGGU_____UUGGGUUUUUUU ..........(((((((((((....))))).))))))((((.....(((((((((((..(((((.....)))))..)))))...........)))..))).......))))...... (-28.96 = -30.12 + 1.16)

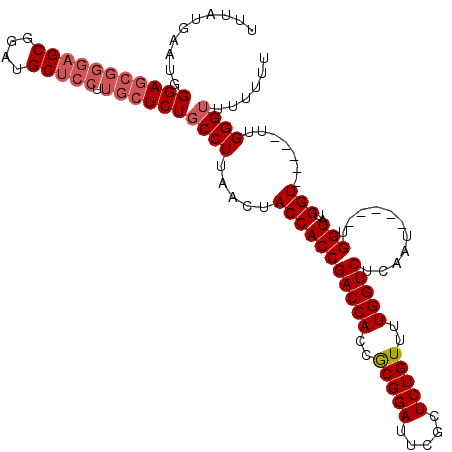
| Location | 8,709,998 – 8,710,105 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 90.76 |
| Mean single sequence MFE | -30.34 |
| Consensus MFE | -23.06 |
| Energy contribution | -23.86 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8709998 107 - 20766785 AAAAAAACCCAA-----ACCAUACCA-----AUUGAGACCAAAACAGAGCGAAUCCGCGGUGGUCGGUGGUAGUUGAGGCAGAGCAAGGAGCAUCCGCUCCCGCUCCCAUUCAUAAA .......(((((-----....(((((-----.....(((((.......(((....)))..)))))..))))).))).))..((((..(((((....))))).))))........... ( -32.90) >DroSec_CAF1 16178 106 - 1 AA-AAAACCCAA-----ACCAUACCA-----AUUGAGACCAAAACAGAGCGAAUCCGCGGUGGUCGGUGGUAGUUAAGGCAGAGCAAGGAGCAUCCGCUCCCGCUCCCAUUCAUAAA ..-....((..(-----((..(((((-----.....(((((.......(((....)))..)))))..))))))))..))..((((..(((((....))))).))))........... ( -30.80) >DroSim_CAF1 19159 107 - 1 AAAAAAACCCAA-----ACCAUACCA-----AUUGAGACCAAAACAGAGCGAAUCCGCGGUGGUCGGUGGUAGUUAAGGCAGAGCAAGGAGCAUCCGCUCCCGCUCCCAUUCAUAAA .......((..(-----((..(((((-----.....(((((.......(((....)))..)))))..))))))))..))..((((..(((((....))))).))))........... ( -30.80) >DroEre_CAF1 16955 106 - 1 AAAAAACCCCAAACCAAACCGAACCA-----AUUGAGACCAAAACAGAGCGAAUCCGUGGUGGUCGGUGGUAGUUAAGGCAGAGCAAGGAGCAUCCGC------UCCCAUUCAUAAA .....(((((.........(((....-----.))).(((((..((.((.....)).))..))))))).)))..........(((...(((((....))------)))..)))..... ( -25.10) >DroYak_CAF1 16946 117 - 1 AAAAGAACCCAAACCAAACCAUACCAUACAUAUUGAGACCAAAACAGAGCGAAUCCGUGGUGGUCGGUGGUAGUUAAGGCAGAGCAAGGAGCAUCCGCUCCCGCUCCCAUUCAUAAA ....(((..............((((((.........(((((..((.((.....)).))..))))).)))))).....((..((((..(((((....))))).)))))).)))..... ( -32.10) >consensus AAAAAAACCCAA_____ACCAUACCA_____AUUGAGACCAAAACAGAGCGAAUCCGCGGUGGUCGGUGGUAGUUAAGGCAGAGCAAGGAGCAUCCGCUCCCGCUCCCAUUCAUAAA .................(((((..((.......)).(((((.......(((....)))..))))).)))))......((..((((..(((((....))))).))))))......... (-23.06 = -23.86 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:43 2006