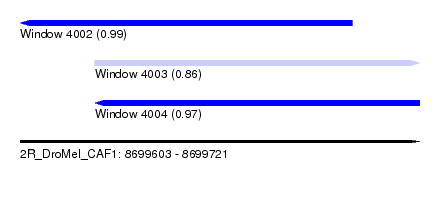
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,699,603 – 8,699,721 |
| Length | 118 |
| Max. P | 0.994074 |

| Location | 8,699,603 – 8,699,701 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 65.75 |
| Mean single sequence MFE | -31.13 |
| Consensus MFE | -8.16 |
| Energy contribution | -6.99 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.45 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.26 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8699603 98 - 20766785 CACAACACAA-ACACAAUCCGUUGCCAGCUCCUCCUGCUCCGCCCCAAGCGAUGGAGGAGGAGGAGGAGUCUACACAAAGUUGG-------------CACCAGUUUCAUCCU ..........-.........(.((((((((((((((.((((...(((.....))).)))).)))))))((....))...)))))-------------)).)........... ( -33.60) >DroPse_CAF1 9890 93 - 1 C----CAAAG-ACACUAUCCGUUGGCAACACUGCACUAUCCCUCCCC---UU-AGAAGGGGUGGGGGAUA--CUCCAACUUUGGCCA---A----UUCACAAAUUUUACUU- (----(((((-....(((((....(((....))).....(((.((((---(.-...))))).))))))))--......))))))...---.----................- ( -27.80) >DroSec_CAF1 6010 104 - 1 CAGAACACAA-ACACAAUCCGUUGCCAGCUCCUCCUGCUCCCCCGCAAGCAA---AGGAGAAGGAGGAGUCUGUCCAACGUUGGCCACACA----CACACCAGUUUCAUCCU ..(((.((..-........(((((.(((((((((((.((((..(....)...---.)))).)))))))).)))..))))).(((.......----....))))))))..... ( -33.60) >DroSim_CAF1 6053 104 - 1 CUCAACACAA-ACACAAUCCGUUGGCAGCUCCUCCUGCUCCCCCGCAAACAA---AGGAGGAGGAGGAGUUUGUCCAACGUUGGCCACACA----CACACCAGUUUCAUCCU ........((-((......(((((((((((((((((.((((...........---.)))).)))))))).))).)))))).(((.......----....)))))))...... ( -35.20) >DroYak_CAF1 6334 106 - 1 CACCACACAAAACCCAAUCCGUUCCCAGCUCC---UGCUCCUUCGCAAAAG---GUGUAGGAGGAGGAGUCUGACCAACGUUGGCCACACACACACACACCAGUUUCAUCCU .............((((..((((..(((((((---(.(((((.(((.....---))).))))).))))).)))...))))))))............................ ( -29.40) >DroPer_CAF1 10427 93 - 1 C----CAAAG-ACACUAUCCGUUGGCAACACUGCACUAUCCCUCCCC---UU-AGAAGGGGUGGGGGGUA--CUCCAACUUUGGCCA---A----UUCACAAAUUUUACUU- (----(((((-....(((((....(((....))).....(((.((((---(.-...))))).))))))))--......))))))...---.----................- ( -27.20) >consensus CACAACACAA_ACACAAUCCGUUGCCAGCUCCUCCUGCUCCCCCCCAA_CAA__GAGGAGGAGGAGGAGUCUGUCCAACGUUGGCCA___A____CACACCAGUUUCAUCCU .........................((((.......(((((.((..................)).))))).........))))............................. ( -8.16 = -6.99 + -1.16)

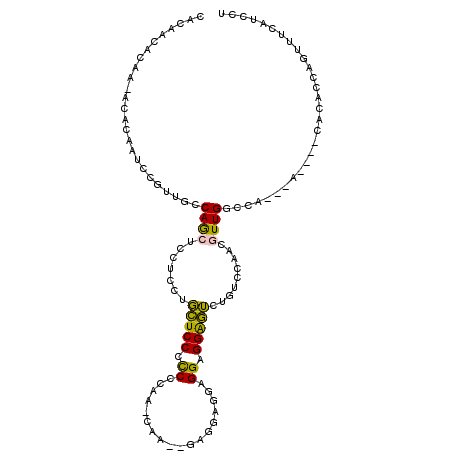
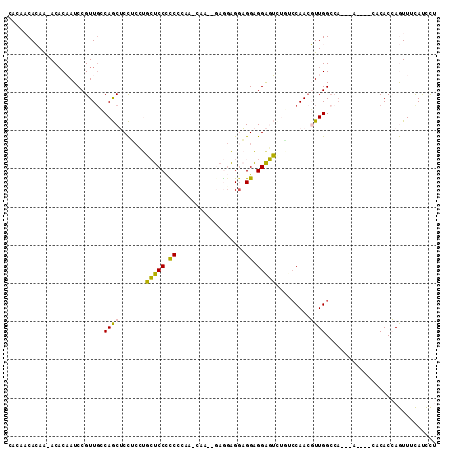
| Location | 8,699,625 – 8,699,721 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 67.26 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -7.27 |
| Energy contribution | -6.97 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.54 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.21 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856107 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
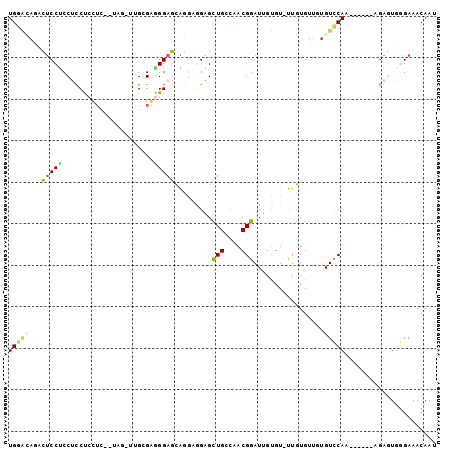
>2R_DroMel_CAF1 8699625 96 + 20766785 UGUGUAGACUCCUCCUCCUCCUCCAUCGCUUGGGGCGGAGCAGGAGGAGCUGGCAACGGAUUGUGU-UUGUGUUGUGUCCAA------AGAGUGGGAAACAAU (((.(((.((((((((.((((((((.....))))..)))).))))))))))))))((((((...))-))))(((((.(((..------......))).))))) ( -35.00) >DroPse_CAF1 9917 85 + 1 UGGAG--UAUCCCCCACCCCUUCU-AA---GGGGAGGGAUAGUGCAGUGUUGCCAACGGAUAGUGU-CUUUG----GUUCAC------AGA-UAAGAAACAAG ((.(.--((((((...(((((...-.)---)))).)))))).).))(((..(((((.((((...))-)))))----)).)))------...-........... ( -28.70) >DroSec_CAF1 6041 93 + 1 UGGACAGACUCCUCCUUCUCCU---UUGCUUGCGGGGGAGCAGGAGGAGCUGGCAACGGAUUGUGU-UUGUGUUCUGUCCAA------AGAGUGGGAAACAAU ((((((((((((((((.(((((---(((....)))))))).))))))))(((....))).......-......)))))))).------......(....)... ( -44.70) >DroSim_CAF1 6084 93 + 1 UGGACAAACUCCUCCUCCUCCU---UUGUUUGCGGGGGAGCAGGAGGAGCUGCCAACGGAUUGUGU-UUGUGUUGAGUCCAA------AGAGUGGGAAACAAU (((((...((((((((.(((((---(((....)))))))).))))))))....(((((.(......-.).))))).))))).------......(....)... ( -39.10) >DroYak_CAF1 6369 97 + 1 UGGUCAGACUCCUCCUCCUACAC---CUUUUGCGAAGGAGCA---GGAGCUGGGAACGGAUUGGGUUUUGUGUGGUGUCCAGUGUGGUGGAGUGGGAAACAAU .........(((..((((..(((---(((((((......)))---))))(((((..((.((........)).))...))))).)))..))))..)))...... ( -30.40) >DroPer_CAF1 10454 85 + 1 UGGAG--UACCCCCCACCCCUUCU-AA---GGGGAGGGAUAGUGCAGUGUUGCCAACGGAUAGUGU-CUUUG----GUUCAC------AGA-UAAGAAACAAG ....(--(((..(((.(((((...-.)---)))).)))...)))).(((..(((((.((((...))-)))))----)).)))------...-........... ( -27.70) >consensus UGGACAGACUCCUCCUCCUCCUC__UAG_UUGCGAGGGAGCAGGAGGAGCUGCCAACGGAUUGUGU_UUGUGUUGUGUCCAA______AGAGUGGGAAACAAU (((((...(((((......................))))).........(((....))).................)))))...................... ( -7.27 = -6.97 + -0.30)

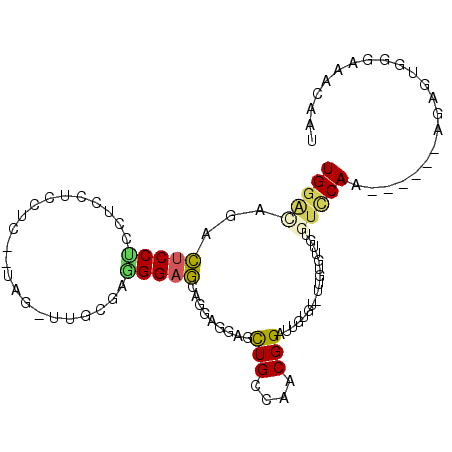
| Location | 8,699,625 – 8,699,721 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 67.26 |
| Mean single sequence MFE | -28.91 |
| Consensus MFE | -6.64 |
| Energy contribution | -5.37 |
| Covariance contribution | -1.27 |
| Combinations/Pair | 1.57 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.23 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
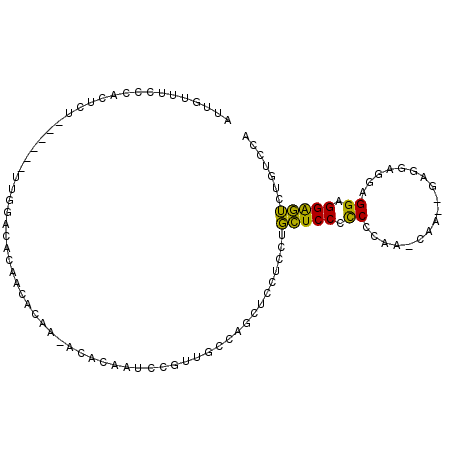
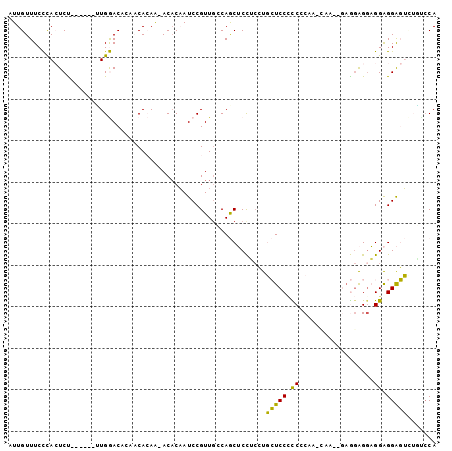
>2R_DroMel_CAF1 8699625 96 - 20766785 AUUGUUUCCCACUCU------UUGGACACAACACAA-ACACAAUCCGUUGCCAGCUCCUCCUGCUCCGCCCCAAGCGAUGGAGGAGGAGGAGGAGUCUACACA .((((.(((......------..))).)))).....-................(((((((((.((((...(((.....))).)))).)))))))))....... ( -32.10) >DroPse_CAF1 9917 85 - 1 CUUGUUUCUUA-UCU------GUGAAC----CAAAG-ACACUAUCCGUUGGCAACACUGCACUAUCCCUCCCC---UU-AGAAGGGGUGGGGGAUA--CUCCA .(((.(((...-...------..))).----)))..-....(((((....(((....))).....(((.((((---(.-...))))).))))))))--..... ( -24.40) >DroSec_CAF1 6041 93 - 1 AUUGUUUCCCACUCU------UUGGACAGAACACAA-ACACAAUCCGUUGCCAGCUCCUCCUGCUCCCCCGCAAGCAA---AGGAGAAGGAGGAGUCUGUCCA ...............------.((((((((.....(-((.......))).....((((((((.((((..(....)...---.)))).)))))))))))))))) ( -33.20) >DroSim_CAF1 6084 93 - 1 AUUGUUUCCCACUCU------UUGGACUCAACACAA-ACACAAUCCGUUGGCAGCUCCUCCUGCUCCCCCGCAAACAA---AGGAGGAGGAGGAGUUUGUCCA ...............------.((((((((((....-.........))))).((((((((((.((((...........---.)))).)))))))))).))))) ( -31.62) >DroYak_CAF1 6369 97 - 1 AUUGUUUCCCACUCCACCACACUGGACACCACACAAAACCCAAUCCGUUCCCAGCUCC---UGCUCCUUCGCAAAAG---GUGUAGGAGGAGGAGUCUGACCA .((((.......((((......))))......))))...............(((((((---(.(((((.(((.....---))).))))).))))).))).... ( -28.32) >DroPer_CAF1 10454 85 - 1 CUUGUUUCUUA-UCU------GUGAAC----CAAAG-ACACUAUCCGUUGGCAACACUGCACUAUCCCUCCCC---UU-AGAAGGGGUGGGGGGUA--CUCCA .(((.(((...-...------..))).----)))..-....(((((....(((....))).....(((.((((---(.-...))))).))))))))--..... ( -23.80) >consensus AUUGUUUCCCACUCU______UUGGACACAACACAA_ACACAAUCCGUUGCCAGCUCCUCCUGCUCCCCCCCAA_CAA__GAGGAGGAGGAGGAGUCUGUCCA ..............................................................(((((.((..................)).)))))....... ( -6.64 = -5.37 + -1.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:40 2006