
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
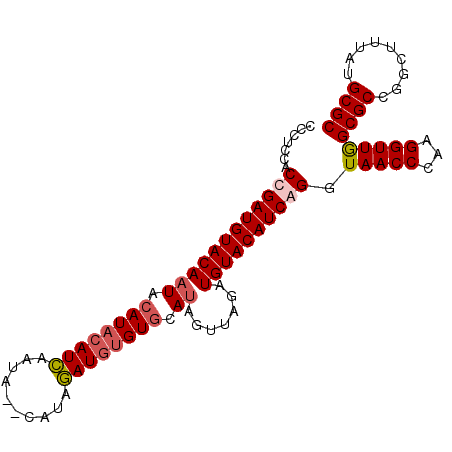
| Location | 8,687,521 – 8,687,613 |
| Length | 92 |
| Max. P | 0.968009 |

| Location | 8,687,521 – 8,687,613 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -21.80 |
| Energy contribution | -22.73 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
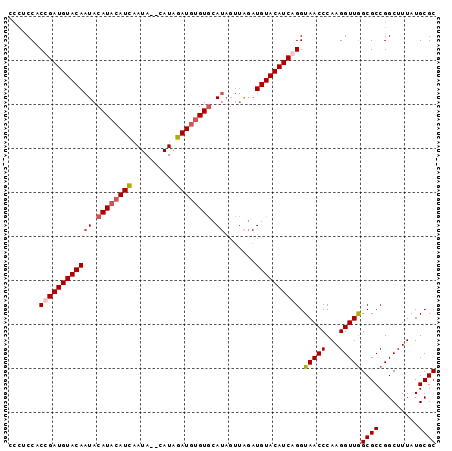
>2R_DroMel_CAF1 8687521 92 + 20766785 ---UCCCCGGAUGUACAAUACAUACAUCAAUA--CAUAGAUGUGUGCAUAGUUAGAUGUACAUCAGGUAACCCAAGGUUGGCGCCGGCUUUAUGCGC ---...(((((((((((((.((((((((....--....)))))))).)).......)))))))).(.(((((...))))).)..))).......... ( -25.71) >DroSec_CAF1 107 95 + 1 CCCUCCACCGAUGUACAAUACAUACAUCAAUA--CAUAGAUGUGUGCAUAGUUAGAUGUACAUCGGGUAACCCAAGGUUAGCGCCGGCUUUAUGCGC .......((((((((((((.((((((((....--....)))))))).)).......)))))))))).(((((...)))))((((.........)))) ( -29.81) >DroSim_CAF1 107 95 + 1 CCCACCACCGAUGUACAAUACAUACAUCAAUA--CAUAGAUGUGUGCAUAGUUAGAUGUACAUCGGGUAACCCAAGGUUGGCGCCGGCUUUAUGCGC .......((((((((((((.((((((((....--....)))))))).)).......)))))))))).......(((((((....)))))))...... ( -29.31) >DroYak_CAF1 107 90 + 1 CCCUCCACGGAUGUACA-UACAUAAAUGUAUAAAAAUGUAU--GUACAU----GUAUGUACAUCAGGUAACCCAAGGUUGGCGCCGGCUUUAUGCGC .......((((((((((-(((((..(((((((....)))))--))..))----))))))))))).(.(((((...))))).)..))((.....)).. ( -28.30) >consensus CCCUCCACCGAUGUACAAUACAUACAUCAAUA__CAUAGAUGUGUGCAUAGUUAGAUGUACAUCAGGUAACCCAAGGUUGGCGCCGGCUUUAUGCGC .......((((((((((((.((((((((..........)))))))).)).......)))))))))).(((((...)))))((((.........)))) (-21.80 = -22.73 + 0.94)

| Location | 8,687,521 – 8,687,613 |
|---|---|
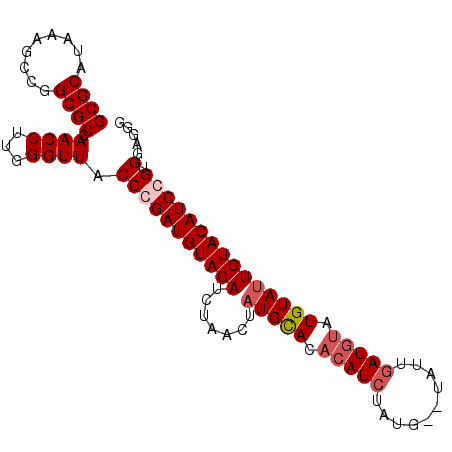
| Length | 92 |
| Sequences | 4 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 86.11 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -21.55 |
| Energy contribution | -22.86 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
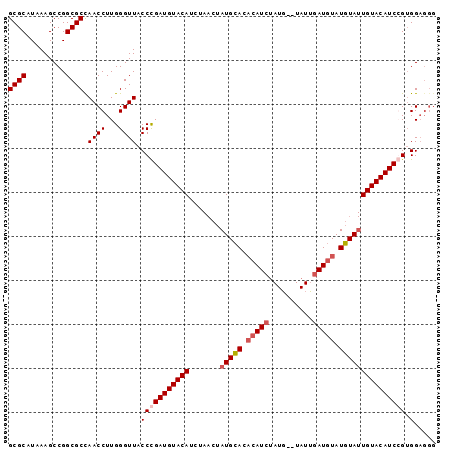
>2R_DroMel_CAF1 8687521 92 - 20766785 GCGCAUAAAGCCGGCGCCAACCUUGGGUUACCUGAUGUACAUCUAACUAUGCACACAUCUAUG--UAUUGAUGUAUGUAUUGUACAUCCGGGGA--- ((((.........))))...(((((((((((...(((((((((.....(((((........))--))).)))))))))...))).)))))))).--- ( -28.60) >DroSec_CAF1 107 95 - 1 GCGCAUAAAGCCGGCGCUAACCUUGGGUUACCCGAUGUACAUCUAACUAUGCACACAUCUAUG--UAUUGAUGUAUGUAUUGUACAUCGGUGGAGGG ((((.........))))...((((.(.....((((((((((.......(((((.(((((....--....))))).)))))))))))))))).)))). ( -32.51) >DroSim_CAF1 107 95 - 1 GCGCAUAAAGCCGGCGCCAACCUUGGGUUACCCGAUGUACAUCUAACUAUGCACACAUCUAUG--UAUUGAUGUAUGUAUUGUACAUCGGUGGUGGG ..........(((.((((......(((...)))((((((((.......(((((.(((((....--....))))).))))))))))))))))).))). ( -33.41) >DroYak_CAF1 107 90 - 1 GCGCAUAAAGCCGGCGCCAACCUUGGGUUACCUGAUGUACAUAC----AUGUAC--AUACAUUUUUAUACAUUUAUGUA-UGUACAUCCGUGGAGGG ((((.........))))...((((((....)).(((((((((((----(((...--.................))))))-))))))))....)))). ( -27.65) >consensus GCGCAUAAAGCCGGCGCCAACCUUGGGUUACCCGAUGUACAUCUAACUAUGCACACAUCUAUG__UAUUGAUGUAUGUAUUGUACAUCCGUGGAGGG ((((.........)))).((((...)))).(((((((((((.......(((((.(((((..........))))).))))))))))))))).)..... (-21.55 = -22.86 + 1.31)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:35 2006