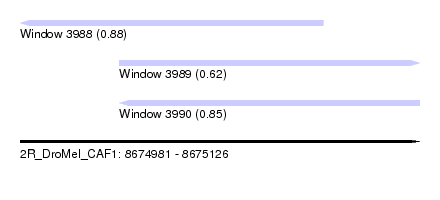
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,674,981 – 8,675,126 |
| Length | 145 |
| Max. P | 0.883622 |

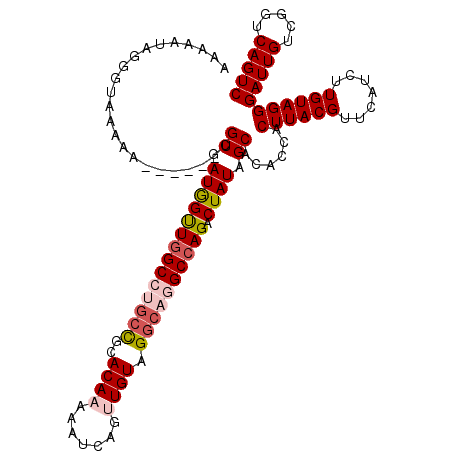
| Location | 8,674,981 – 8,675,091 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 88.73 |
| Mean single sequence MFE | -32.70 |
| Consensus MFE | -26.90 |
| Energy contribution | -28.14 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8674981 110 - 20766785 AAAAAUAGGGUAAAAA------CGUGAUGGUUGGCCUGCCGCACAAAAAUCAGUUGUAGGCAGGCCAGACUAUAGCACAACACUUACGUUCAUCUUGUAGGGAUUGUCGGUCAGUC ................------.((.((((((((((((((..((((.......)))).))))))))).))))).))((((..((((((.......))))))..))))......... ( -35.50) >DroSec_CAF1 329 110 - 1 AAAAAUAGUGUAAAAA------CGUGAUGGUUUGCCUGCUGCACAAAAAUCAGUUGUAGGCAGGCCAGACUAUAGCACACCACUUACGUUCAUCUUGUAGGGAUUGUCGGUCAGUC .......((((.....------.((..((((((((((((.((..........)).)))))))))))).))....))))....((((((.......))))))(((((.....))))) ( -31.20) >DroSim_CAF1 59 110 - 1 AAAAAUAGUGUAAAAA------CGUGAUGGUUGGCCUGCCGCACAAAAAUCAGUUGUAGGCAGGCCAGACUAUAGCACACCACUUACGUUCAUCUUGUAGGGAUUGUCGGUCAGUC .......((((.....------.((.((((((((((((((..((((.......)))).))))))))).))))).))))))..((((((.......))))))(((((.....))))) ( -35.10) >DroEre_CAF1 59 114 - 1 AGAAAUAGGGCAAAAAC--GAGCGUGAUAGCUGGCCGGCCGUACAAAAAUCAGCUGUAGGCAAGCCAGACUAUAGCAUACCACUUACGUUCAUCUUGUAGGGAUUGUCGGUCAGUC ........(((((....--..((...(((((((((..(((.((((.........)))))))..))))).)))).))......((((((.......))))))..)))))........ ( -32.90) >DroYak_CAF1 59 112 - 1 AGAAAUAUGCCAAAAUCAAAAGCGUGAUAGUUGGC----CGCACAAAAAUCAGCUGUAGGCAGGCCAGACUAUAGCAUGCCACUUACGUUCAUCUUGUAGGGAUUGUCGGUCAGUC ........(((..((((....((((((((((((((----((((((.........)))..)).))))).)))))..)))))..((((((.......))))))))))...)))..... ( -28.80) >consensus AAAAAUAGGGUAAAAA______CGUGAUGGUUGGCCUGCCGCACAAAAAUCAGUUGUAGGCAGGCCAGACUAUAGCACACCACUUACGUUCAUCUUGUAGGGAUUGUCGGUCAGUC .......................((.((((((((((((((..((((.......)))).)))))))))).)))).))......((((((.......))))))(((((.....))))) (-26.90 = -28.14 + 1.24)

| Location | 8,675,017 – 8,675,126 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 85.97 |
| Mean single sequence MFE | -27.94 |
| Consensus MFE | -19.36 |
| Energy contribution | -20.88 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8675017 109 + 20766785 UUGUGCUAUAGUCUGGCCUGCCUACAACUGAUUUUUGUGCGGCAGGCCAACCAUCACG------UUUUUACCCUAUUUUUGGCAGCAGUUUUGUUAUCUCACAGCACGCACUUCA ..(((((...(..((((((((((((((.......))))).)))))))))..)......------...............((((((.....))))))......)))))........ ( -32.00) >DroSec_CAF1 365 109 + 1 GUGUGCUAUAGUCUGGCCUGCCUACAACUGAUUUUUGUGCAGCAGGCAAACCAUCACG------UUUUUACACUAUUUUUGGCAGCAGUUUUGUUAUCUCACAGCACUCACUCCA (.((((((((((...((((((.(((((.......)))))..))))))((((......)------)))....)))))...((((((.....))))))......))))).)...... ( -24.00) >DroSim_CAF1 95 109 + 1 GUGUGCUAUAGUCUGGCCUGCCUACAACUGAUUUUUGUGCGGCAGGCCAACCAUCACG------UUUUUACACUAUUUUUGGCAGCAGUUUUGUUAUCUCACAGCACUCACUCCA (.((((((((((.((((((((((((((.......))))).)))))))))........(------(....)))))))...((((((.....))))))......))))).)...... ( -32.70) >DroEre_CAF1 95 113 + 1 GUAUGCUAUAGUCUGGCUUGCCUACAGCUGAUUUUUGUACGGCCGGCCAGCUAUCACGCUC--GUUUUUGCCCUAUUUCUGUCAGCAGUUUGGUUAUCUCAUAUCACUCACUCCA ...((((((((.((((((.((((((((.......))))).))).))))))))))...((..--......))............))))(..((((........))))..)...... ( -27.60) >DroYak_CAF1 95 110 + 1 GCAUGCUAUAGUCUGGCCUGCCUACAGCUGAUUUUUGUGCG----GCCAACUAUCACGCUUUUGAUUUUGGCAUAUUUCUGGCAGCAGUUUUGUUAUCUCAUAGC-CUCACUCCA ((.(((((((((.(((((.((..((((.......)))))))----))))))))....(((.........))).......))))))).(((.((......)).)))-......... ( -23.40) >consensus GUGUGCUAUAGUCUGGCCUGCCUACAACUGAUUUUUGUGCGGCAGGCCAACCAUCACG______UUUUUACCCUAUUUUUGGCAGCAGUUUUGUUAUCUCACAGCACUCACUCCA ..(((((...((.((((((((((((((.......))))).))))))))))).............................(..(((......)))..)....)))))........ (-19.36 = -20.88 + 1.52)

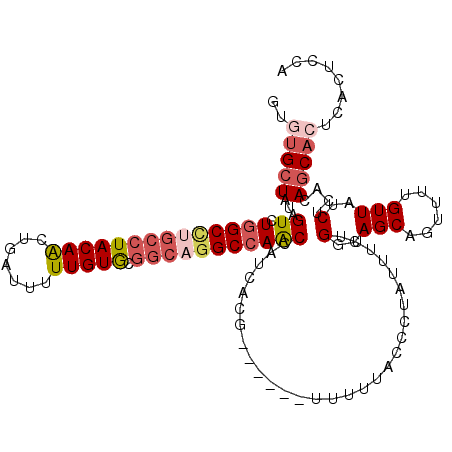
| Location | 8,675,017 – 8,675,126 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 85.97 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -21.82 |
| Energy contribution | -24.46 |
| Covariance contribution | 2.64 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8675017 109 - 20766785 UGAAGUGCGUGCUGUGAGAUAACAAAACUGCUGCCAAAAAUAGGGUAAAAA------CGUGAUGGUUGGCCUGCCGCACAAAAAUCAGUUGUAGGCAGGCCAGACUAUAGCACAA ....((((((..(((......)))..))...((((........))))....------....((((((((((((((..((((.......)))).))))))))).))))).)))).. ( -37.20) >DroSec_CAF1 365 109 - 1 UGGAGUGAGUGCUGUGAGAUAACAAAACUGCUGCCAAAAAUAGUGUAAAAA------CGUGAUGGUUUGCCUGCUGCACAAAAAUCAGUUGUAGGCAGGCCAGACUAUAGCACAC ((((((.(((..(((......)))..)))))).)))......((((.....------.((..((((((((((((.((..........)).)))))))))))).))....)))).. ( -33.30) >DroSim_CAF1 95 109 - 1 UGGAGUGAGUGCUGUGAGAUAACAAAACUGCUGCCAAAAAUAGUGUAAAAA------CGUGAUGGUUGGCCUGCCGCACAAAAAUCAGUUGUAGGCAGGCCAGACUAUAGCACAC ((((((.(((..(((......)))..)))))).)))......((......)------)(((((((((((((((((..((((.......)))).))))))))).)))))..))).. ( -36.90) >DroEre_CAF1 95 113 - 1 UGGAGUGAGUGAUAUGAGAUAACCAAACUGCUGACAGAAAUAGGGCAAAAAC--GAGCGUGAUAGCUGGCCGGCCGUACAAAAAUCAGCUGUAGGCAAGCCAGACUAUAGCAUAC ....((.(((..(.((.......)).)..))).)).................--..((...(((((((((..(((.((((.........)))))))..))))).)))).)).... ( -27.70) >DroYak_CAF1 95 110 - 1 UGGAGUGAG-GCUAUGAGAUAACAAAACUGCUGCCAGAAAUAUGCCAAAAUCAAAAGCGUGAUAGUUGGC----CGCACAAAAAUCAGCUGUAGGCAGGCCAGACUAUAGCAUGC ....(((.(-((((.............(((....)))...(((((...........))))).....))))----).)))........(((((((..........))))))).... ( -24.00) >consensus UGGAGUGAGUGCUGUGAGAUAACAAAACUGCUGCCAAAAAUAGGGUAAAAA______CGUGAUGGUUGGCCUGCCGCACAAAAAUCAGUUGUAGGCAGGCCAGACUAUAGCACAC ((((((.(((..(((......)))..)))))).)))......................((.((((((((((((((..((((.......)))).)))))))))).)))).)).... (-21.82 = -24.46 + 2.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:27 2006