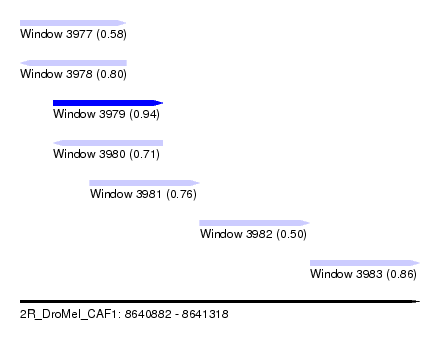
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,640,882 – 8,641,318 |
| Length | 436 |
| Max. P | 0.943390 |

| Location | 8,640,882 – 8,640,998 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 88.17 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -21.14 |
| Energy contribution | -21.82 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8640882 116 + 20766785 UUCCUAUCCAGGGCUGCCUCUCCAGAAGCCAUGCGAGAUGGGUACUAUGCAACCCAUACACAUGCUACACCCUGGCUUAUAGGAUUCCUCUUCGGAUACUUCCUGCAUCUGAACAG .(((((((((((((((......))).(((.(((.(..((((((........)))))).).))))))...))))))...))))))...((.(((((((.(.....).))))))).)) ( -36.70) >DroSec_CAF1 21051 115 + 1 -UUCUAUCCAGGACUGCCUCUCCAGAAGCCAUGCGAGAUGGGUACUAUGCAACCCAUACUCAUGCUACACCCUGGCUGAUAGGAUUCCUCUUCGGAUACUUCCUGCAUCUGAACAG -(((((((((((...((.((....)).)).....(((((((((........)))))).))).........)))))...))))))...((.(((((((.(.....).))))))).)) ( -31.20) >DroSim_CAF1 24728 115 + 1 -UUCUACCCAGGACUGCCUCGUCAGAAGCCAUGCGAGAUGGGUACUAUGCAACCCAUACUCAUGCUACACCCUGGCUGAUAGGAUUCCUCUUCGGAUACUUCCUGCAUCUGAACAG -........(((....)))..((((((((((.(((((((((((........)))))).)))..)).......)))))(.(((((....((....))....)))))).))))).... ( -31.00) >DroYak_CAF1 21026 115 + 1 -UUUUCUCCAGAAAUGCAACUCCAGGAGCAACGGAAGCUGCAUACUAUGCAACCCAUACGCACGCUACACCUUGGCUAAUAGGAUUCCUCUUCGGAUACUUCCUGCAUCUGAGCAA -.......((((..(((....(((((.(((.(....).)))......(((.........)))........))))).....((((....((....))....)))))))))))..... ( -25.80) >consensus _UUCUAUCCAGGACUGCCUCUCCAGAAGCCAUGCGAGAUGGGUACUAUGCAACCCAUACUCAUGCUACACCCUGGCUGAUAGGAUUCCUCUUCGGAUACUUCCUGCAUCUGAACAG ........((((..(((....((((.(((.(((....((((((........))))))...)))))).....)))).....((((.(((.....)))....)))))))))))..... (-21.14 = -21.82 + 0.69)

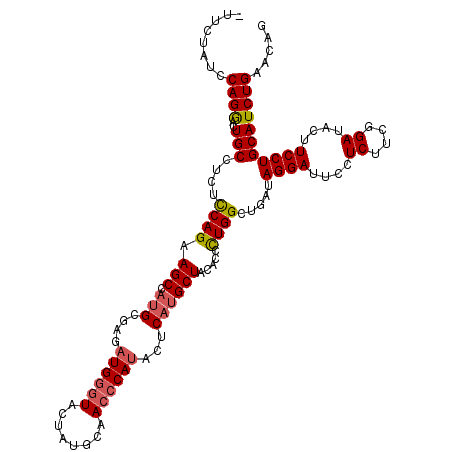
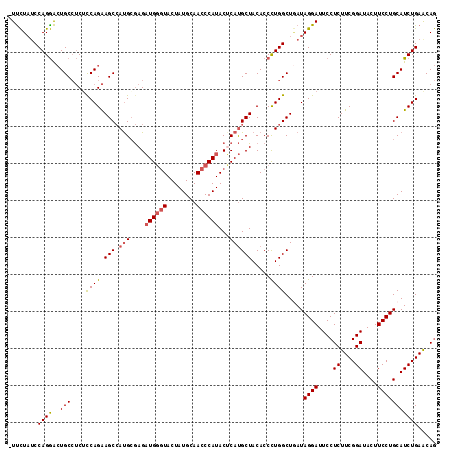
| Location | 8,640,882 – 8,640,998 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 88.17 |
| Mean single sequence MFE | -41.07 |
| Consensus MFE | -28.21 |
| Energy contribution | -29.40 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8640882 116 - 20766785 CUGUUCAGAUGCAGGAAGUAUCCGAAGAGGAAUCCUAUAAGCCAGGGUGUAGCAUGUGUAUGGGUUGCAUAGUACCCAUCUCGCAUGGCUUCUGGAGAGGCAGCCCUGGAUAGGAA ((.(((.(((((.....))))).))).))...((((((...(((((((....((((((.((((((........))))))..))))))(((((....))))).))))))))))))). ( -51.30) >DroSec_CAF1 21051 115 - 1 CUGUUCAGAUGCAGGAAGUAUCCGAAGAGGAAUCCUAUCAGCCAGGGUGUAGCAUGAGUAUGGGUUGCAUAGUACCCAUCUCGCAUGGCUUCUGGAGAGGCAGUCCUGGAUAGAA- ((((((((.(((((((....(((.....))).)))).....((((((.((....((((.((((((........))))))))))....))))))))....)))...))))))))..- ( -39.50) >DroSim_CAF1 24728 115 - 1 CUGUUCAGAUGCAGGAAGUAUCCGAAGAGGAAUCCUAUCAGCCAGGGUGUAGCAUGAGUAUGGGUUGCAUAGUACCCAUCUCGCAUGGCUUCUGACGAGGCAGUCCUGGGUAGAA- ((.(((.(((((.....))))).))).)).....((((...((((((....((..(((.((((((........)))))))))))...(((((....)))))..))))))))))..- ( -38.90) >DroYak_CAF1 21026 115 - 1 UUGCUCAGAUGCAGGAAGUAUCCGAAGAGGAAUCCUAUUAGCCAAGGUGUAGCGUGCGUAUGGGUUGCAUAGUAUGCAGCUUCCGUUGCUCCUGGAGUUGCAUUUCUGGAGAAAA- ...(((.(((((((((....(((.....))).)))).....(((.((.((((((.(.....(((((((((...)))))))))))))))).)))))....)))))....)))....- ( -34.60) >consensus CUGUUCAGAUGCAGGAAGUAUCCGAAGAGGAAUCCUAUCAGCCAGGGUGUAGCAUGAGUAUGGGUUGCAUAGUACCCAUCUCGCAUGGCUUCUGGAGAGGCAGUCCUGGAUAGAA_ ((((((((.(((((((....(((.....))).)))).....((((((.((....((((.((((((........))))))))))....))))))))....)))...))))))))... (-28.21 = -29.40 + 1.19)

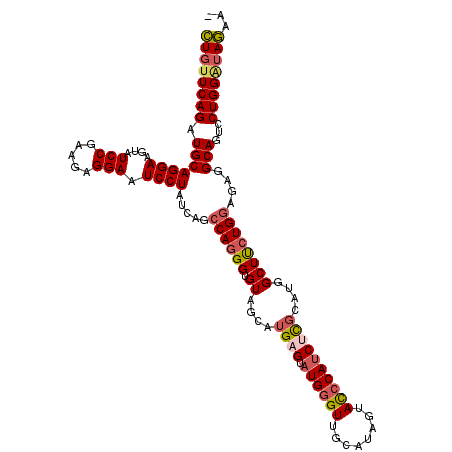
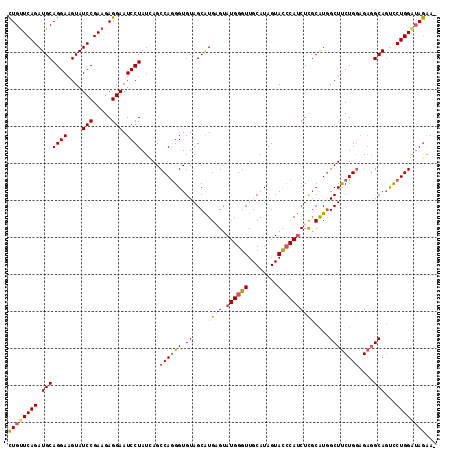
| Location | 8,640,918 – 8,641,038 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.06 |
| Mean single sequence MFE | -43.65 |
| Consensus MFE | -36.81 |
| Energy contribution | -38.62 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8640918 120 + 20766785 GAUGGGUACUAUGCAACCCAUACACAUGCUACACCCUGGCUUAUAGGAUUCCUCUUCGGAUACUUCCUGCAUCUGAACAGAGGGAAGAAGUUCCAGCUUAGCUGGGUGUCCGUGUGGUCA .((((((........)))))).((((((..((((((.((((....((((((((((((((((.(.....).))))))..)))))))......))).....)))))))))).)))))).... ( -44.80) >DroSec_CAF1 21086 120 + 1 GAUGGGUACUAUGCAACCCAUACUCAUGCUACACCCUGGCUGAUAGGAUUCCUCUUCGGAUACUUCCUGCAUCUGAACAGAGGGAAGAAGUUCCAGCUCAGCUGGGUAGCCGUGUGGUCA .((((((........))))))......(((((((((..(((((..((((((((((((((((.(.....).))))))..)))))))......)))...)))))..)).....))))))).. ( -43.80) >DroSim_CAF1 24763 120 + 1 GAUGGGUACUAUGCAACCCAUACUCAUGCUACACCCUGGCUGAUAGGAUUCCUCUUCGGAUACUUCCUGCAUCUGAACAGGGGGAAAAAGUUCCAGCUCAGCUGGGUGUCCGUGUGGUCA .((((((........))))))..((((((.((((((.((((((..((((((((((((((((.(.....).))))))...))))))).....)))...))))))))))))..))))))... ( -45.20) >DroYak_CAF1 21061 120 + 1 GCUGCAUACUAUGCAACCCAUACGCACGCUACACCUUGGCUAAUAGGAUUCCUCUUCGGAUACUUCCUGCAUCUGAGCAAGGGGAAGAAGUUCCAGCUCAGCUGGGUGUCCGUGUGGUCG ..(((((...)))))..(((((((.(((((.............(((((....((....))....)))))((.((((((...((((.....)))).)))))).))))))).)))))))... ( -40.80) >consensus GAUGGGUACUAUGCAACCCAUACUCAUGCUACACCCUGGCUGAUAGGAUUCCUCUUCGGAUACUUCCUGCAUCUGAACAGAGGGAAGAAGUUCCAGCUCAGCUGGGUGUCCGUGUGGUCA .((((((........)))))).((((((..((((((.((((((..((((((((((((((((.(.....).))))))..)))))))......)))...)))))))))))).)))))).... (-36.81 = -38.62 + 1.81)


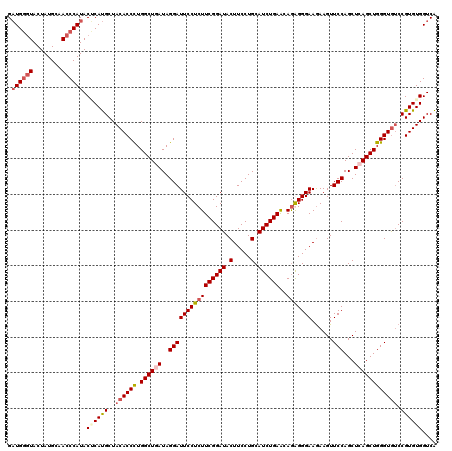
| Location | 8,640,918 – 8,641,038 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.06 |
| Mean single sequence MFE | -41.00 |
| Consensus MFE | -33.25 |
| Energy contribution | -33.38 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.706922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8640918 120 - 20766785 UGACCACACGGACACCCAGCUAAGCUGGAACUUCUUCCCUCUGUUCAGAUGCAGGAAGUAUCCGAAGAGGAAUCCUAUAAGCCAGGGUGUAGCAUGUGUAUGGGUUGCAUAGUACCCAUC .....(((((.((((((.(((.(...((((((((((.(.(((....))).).))))))).(((.....))).)))..).)))..))))))....)))))((((((........)))))). ( -39.80) >DroSec_CAF1 21086 120 - 1 UGACCACACGGCUACCCAGCUGAGCUGGAACUUCUUCCCUCUGUUCAGAUGCAGGAAGUAUCCGAAGAGGAAUCCUAUCAGCCAGGGUGUAGCAUGAGUAUGGGUUGCAUAGUACCCAUC .....((.(((((((((.(((((...((((((((((.(.(((....))).).))))))).(((.....))).)))..)))))...)).))))).)).))((((((........)))))). ( -41.30) >DroSim_CAF1 24763 120 - 1 UGACCACACGGACACCCAGCUGAGCUGGAACUUUUUCCCCCUGUUCAGAUGCAGGAAGUAUCCGAAGAGGAAUCCUAUCAGCCAGGGUGUAGCAUGAGUAUGGGUUGCAUAGUACCCAUC ..((((..(..((((((.(((((...((((....)))).(((.(((.(((((.....))))).))).))).......)))))..)))))).)..)).))((((((........)))))). ( -40.60) >DroYak_CAF1 21061 120 - 1 CGACCACACGGACACCCAGCUGAGCUGGAACUUCUUCCCCUUGCUCAGAUGCAGGAAGUAUCCGAAGAGGAAUCCUAUUAGCCAAGGUGUAGCGUGCGUAUGGGUUGCAUAGUAUGCAGC ...(((((((.(((((..(((((...(((.(((((((....(((......)))(((....))))))))))..)))..)))))...)))))..))))....)))(((((((...))))))) ( -42.30) >consensus UGACCACACGGACACCCAGCUGAGCUGGAACUUCUUCCCCCUGUUCAGAUGCAGGAAGUAUCCGAAGAGGAAUCCUAUCAGCCAGGGUGUAGCAUGAGUAUGGGUUGCAUAGUACCCAUC ...((....))((((((.(((((...(((.(((((((....(((......)))(((....))))))))))..)))..)))))..))))))..........(((((........))))).. (-33.25 = -33.38 + 0.12)

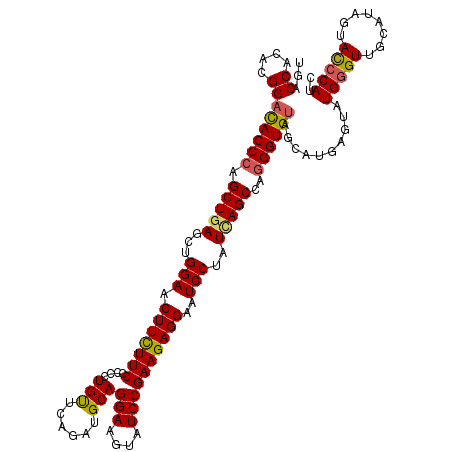
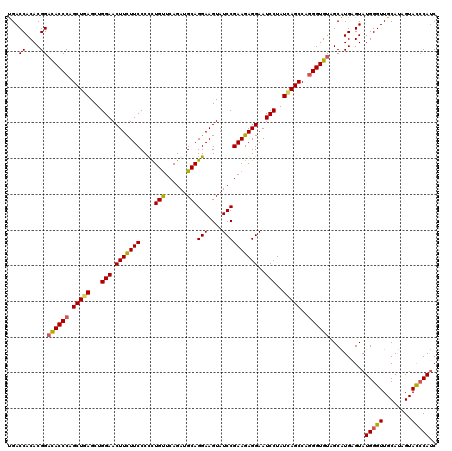
| Location | 8,640,958 – 8,641,078 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -41.27 |
| Consensus MFE | -37.69 |
| Energy contribution | -37.75 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8640958 120 + 20766785 UUAUAGGAUUCCUCUUCGGAUACUUCCUGCAUCUGAACAGAGGGAAGAAGUUCCAGCUUAGCUGGGUGUCCGUGUGGUCAGGAUGGAUCCUCUGCCUGGCCAUGAUCUUUACAUCGAUCU .....((((((((((((((((.(.....).))))))..)))))))......((((((...))))))..)))..(((((((((.(((.....))))))))))))((((........)))). ( -43.10) >DroSec_CAF1 21126 120 + 1 UGAUAGGAUUCCUCUUCGGAUACUUCCUGCAUCUGAACAGAGGGAAGAAGUUCCAGCUCAGCUGGGUAGCCGUGUGGUCAGGAUGGAUCCUCUGUCUGGCCAUGAUCUUCACAUCGAUCU .....((((((((((((((((.(.....).))))))..)))))(((((...((((((...)))))).......((((((((.((((.....))))))))))))..))))).....))))) ( -40.40) >DroSim_CAF1 24803 120 + 1 UGAUAGGAUUCCUCUUCGGAUACUUCCUGCAUCUGAACAGGGGGAAAAAGUUCCAGCUCAGCUGGGUGUCCGUGUGGUCAGGAUGGAUCCUCUGUCUGGCCAUGAUCUUCACAUCGAUCU .....((((((((((((((((.(.....).))))))...))))))).....((((((...))))))..)))..((((((((.((((.....))))))))))))((((........)))). ( -40.30) >DroYak_CAF1 21101 120 + 1 UAAUAGGAUUCCUCUUCGGAUACUUCCUGCAUCUGAGCAAGGGGAAGAAGUUCCAGCUCAGCUGGGUGUCCGUGUGGUCGGGAUGGAUCCUCUGCCUGGCCAUGAUCUUCACAUCGCUCU ...(((((....((....))....))))).....((((...(.(((((...((((((...)))))).......(((((((((.(((.....))))))))))))..))))).)...)))). ( -41.30) >consensus UGAUAGGAUUCCUCUUCGGAUACUUCCUGCAUCUGAACAGAGGGAAGAAGUUCCAGCUCAGCUGGGUGUCCGUGUGGUCAGGAUGGAUCCUCUGCCUGGCCAUGAUCUUCACAUCGAUCU .....((((((((((((((((.(.....).))))))..)))))))......((((((...))))))..)))..(((((((((..(....)....)))))))))((((........)))). (-37.69 = -37.75 + 0.06)

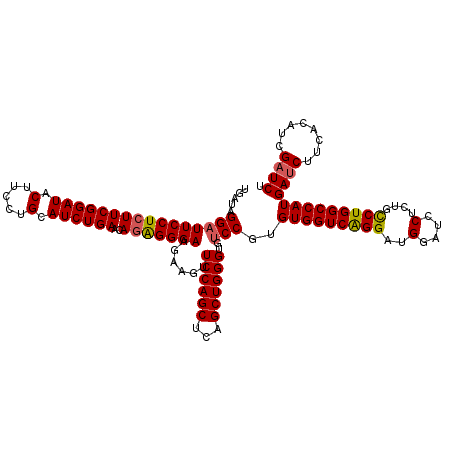
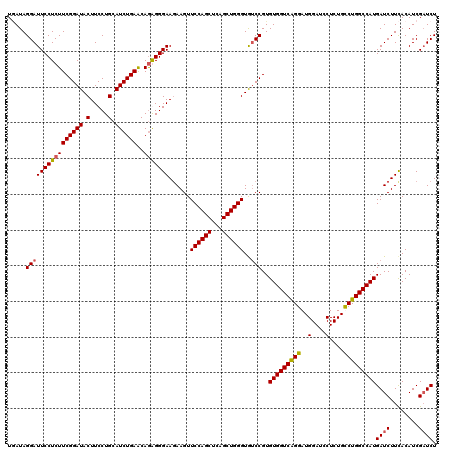
| Location | 8,641,078 – 8,641,198 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.14 |
| Mean single sequence MFE | -41.67 |
| Consensus MFE | -32.39 |
| Energy contribution | -32.57 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8641078 120 + 20766785 UUGCCCUGUAUCCGCCGUCCAAGUGGUCUGAUCCUGACGCGUCCACCUUGGCGGAGUCUUUUUACUACACUUUAACCAGAGUGGGCUGGGCUUUGGCCCUUUGCUGGGUGAUCUUUGCCU (..(((....(((((((.....((((.(((.......)).).))))..)))))))............((((((....))))))(((.((((....))))...))))))..)......... ( -38.90) >DroSec_CAF1 21246 120 + 1 UUGCCCUGUAUCCGCCGUCCAAGUUGUCUGGACCUGACGCGUCCACCUUGGCGGAGUCUUUAUACUACACUUUAACCAGGGUGGGCUGGGCUUUGGCCCUUUGCUGGGUGAUCUUCGCCU (..(((.(((...((((((((.((.(((.......)))))(((((((((((.(((((...........)))))..))))))))))))))))...)))....))).)))..)......... ( -43.40) >DroSim_CAF1 24923 120 + 1 UUGCCCUGUAUCCGCCGUCCAAGUGGUCUGGACCUGACGCGUCCACCUUGGCGGAGUCUUUAUACUACACUUUAACCAGAGUGGGCUGGGCCUUGGCCCUUUGCUGGGUAAUCUUCGCCU ((((((....(((((((.((....)).)((((((....).)))))....))))))............((((((....))))))(((.((((....))))...)))))))))......... ( -43.80) >DroYak_CAF1 21221 120 + 1 UUGCCAUGUAUCCACCGUCCAAGUGGACUGAUUCUUCAGCGUCCACCUUGGCGGAGACUUCUUACUAUACUUUCACGAGAUUGGGCUGGGCGAUGGCCCUUUGCUGGGUGAUCUUCGCCU ..(((....((((.(((.(((((((((((((....)))..))))).))))))))((....))..............).)))..))).(((((((.((((......)))).)...)))))) ( -40.60) >consensus UUGCCCUGUAUCCGCCGUCCAAGUGGUCUGAACCUGACGCGUCCACCUUGGCGGAGUCUUUAUACUACACUUUAACCAGAGUGGGCUGGGCUUUGGCCCUUUGCUGGGUGAUCUUCGCCU ((((((.(((.((.(((.((((((((.(............).))).)))))))).............((((((....))))))))..((((....))))..))).))))))......... (-32.39 = -32.57 + 0.19)

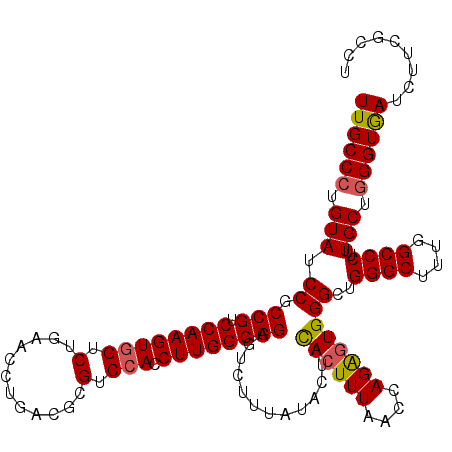
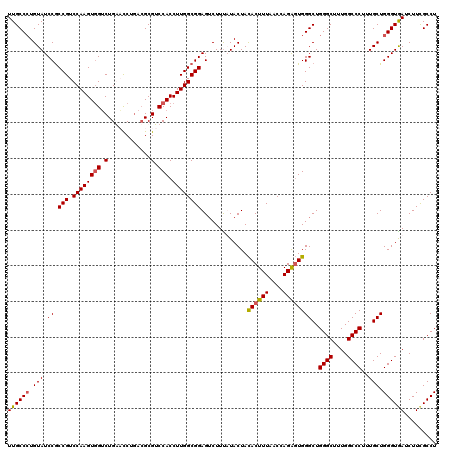
| Location | 8,641,198 – 8,641,318 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -38.08 |
| Consensus MFE | -34.83 |
| Energy contribution | -35.70 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8641198 120 + 20766785 GCGUUCAGGGAUACGGAGGACUGGCCAAUAGUUUCCUCGCCUCUCCGCUAUGGCAACCACUCUCCAGGCUUUCGUAUUCUGUUUUUAUAUGGCACAUGUUCUUCAUGGAGAUGAACGCAC (((((((((((((((((((.((((......(((.((..((......))...)).)))......)))).))))))))))))..............((((.....))))....))))))).. ( -39.00) >DroSec_CAF1 21366 120 + 1 GCAUUCAGGGAUACGGAGGACUUGCCAAUAGUUUCCUCGCCUCUCCGCUAUGGCAACCACUCUCUAGGCUUUCGUAUUCCGUUUUUAUAUGGCACAUGUUCUUCAUGGAGAUGAACGCAC ((.((((((((((((((((..((((((.((((..............))))))))))((........))))))))))))))..............((((.....))))....)))).)).. ( -34.04) >DroSim_CAF1 25043 120 + 1 GCGUUCAGGGAUACGGAGGACUGGCCAAUAGUUUCCUCGCCUCUCCGCUAUGGCAACCACUCUCCAGGCUUUCGUAUUCCGUUUUUAUAUGGCACAUGUUCUUCAUGGAGAUGAACGCAC (((((((((((((((((((.((((......(((.((..((......))...)).)))......)))).))))))))))))..............((((.....))))....))))))).. ( -41.60) >DroYak_CAF1 21341 120 + 1 GCGUUCAGGGACACGGAGGACUGGCCAAUAGUUUCCUCUCCUCUCCGCUAUGGCAACCCCUCUCCAGGCUGUCGUAUUCCGUUUUUAUAUGGCACAUGUUCUUUAUGGAGAUGAACGCCC ((((((((((....((((((..((..........))..))))))..((....))...)))(((((((((((((((((.........))))))))...))).....))))))))))))).. ( -37.70) >consensus GCGUUCAGGGAUACGGAGGACUGGCCAAUAGUUUCCUCGCCUCUCCGCUAUGGCAACCACUCUCCAGGCUUUCGUAUUCCGUUUUUAUAUGGCACAUGUUCUUCAUGGAGAUGAACGCAC (((((((((((((((((((.((((......(((.((..((......))...)).)))......)))).))))))))))))..............((((.....))))....))))))).. (-34.83 = -35.70 + 0.87)

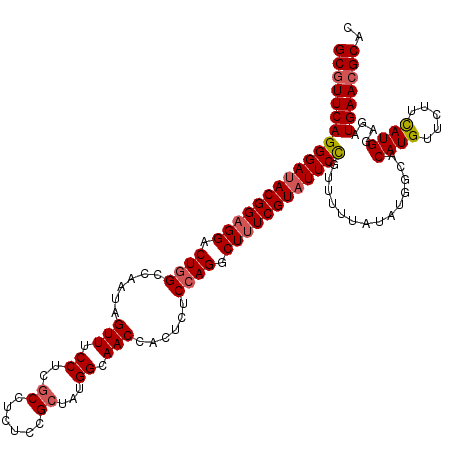
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:20 2006