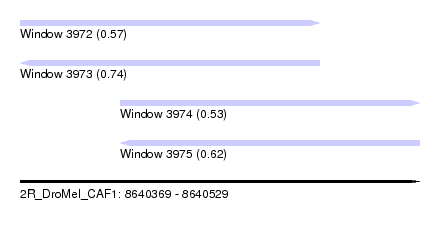
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,640,369 – 8,640,529 |
| Length | 160 |
| Max. P | 0.742968 |

| Location | 8,640,369 – 8,640,489 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -32.49 |
| Energy contribution | -31.42 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.20 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8640369 120 + 20766785 UCCGCAGGACCAAAGGCAAACUUAAUGUGCCAAUGAUGUAUUUACAUCGCUUAAUACGCAUCGCUCCACUUCUGGCAAUGGCCAUAGUGGUCCACAUGAAGCUGAUGCCCUUGAUAUCAA ....((((......((((.........))))...(((((....))))).........(((((((((((((..((((....)))).))))).........))).))))).))))....... ( -31.50) >DroSec_CAF1 20545 120 + 1 UCCGCAGGACCAAAGGCAAACUUAAUGUGCUAAUGAUGUAUUUACAUCGCCUAAUCCGCAUCGCUCCACUUCUGGCAAUGGCCAUAGUGGUCCACAUGAAGCUGAUGCCCUUGAUAUCAA ....((((.....((((..((.....))......(((((....))))))))).....(((((((((((((..((((....)))).))))).........))).))))).))))....... ( -33.10) >DroSim_CAF1 24222 120 + 1 UCCGCAGGACGAAAGGCAAACUUAAUGUGCUAAUGAUGUAUUUACAUCGCCUAAUCCGCAUCGCUCCACUUCUGGCAAUGGCCAUAGUGGUCCACAUGAAGCUGAUGCCCUUGAUAUCAA ....((((.....((((..((.....))......(((((....))))))))).....(((((((((((((..((((....)))).))))).........))).))))).))))....... ( -33.10) >DroYak_CAF1 20516 120 + 1 CCCGCAGAACCAAGGGCAAACUUAAUGUGCCAAUGAUGUAUUUGCACCGCCUCAUCCGCAUCGCUCCGCUCCUGGCGAUGGCCACAGUGGUUCACAUGAAGCUGAUGCCUCUGAUAUCCG ......((((((..(((.........((((.((((...)))).))))...........(((((((........))))))))))....)))))).((.((.((....)).))))....... ( -31.80) >consensus UCCGCAGGACCAAAGGCAAACUUAAUGUGCCAAUGAUGUAUUUACAUCGCCUAAUCCGCAUCGCUCCACUUCUGGCAAUGGCCAUAGUGGUCCACAUGAAGCUGAUGCCCUUGAUAUCAA ....((((.....((((..((.....))......(((((....))))))))).....(((((((((((((..((((....)))).))))).........))).))))).))))....... (-32.49 = -31.42 + -1.06)

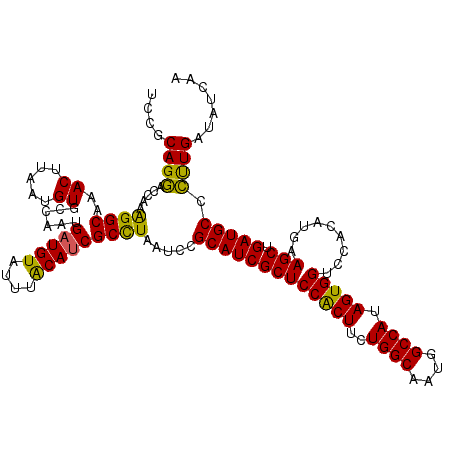
| Location | 8,640,369 – 8,640,489 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -38.90 |
| Consensus MFE | -35.88 |
| Energy contribution | -36.25 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8640369 120 - 20766785 UUGAUAUCAAGGGCAUCAGCUUCAUGUGGACCACUAUGGCCAUUGCCAGAAGUGGAGCGAUGCGUAUUAAGCGAUGUAAAUACAUCAUUGGCACAUUAAGUUUGCCUUUGGUCCUGCGGA .....((((((((((..(((((.(((((..(((((.((((....))))..)))))..((((((.......))(((((....))))))))).))))).)))))))))))))))........ ( -42.30) >DroSec_CAF1 20545 120 - 1 UUGAUAUCAAGGGCAUCAGCUUCAUGUGGACCACUAUGGCCAUUGCCAGAAGUGGAGCGAUGCGGAUUAGGCGAUGUAAAUACAUCAUUAGCACAUUAAGUUUGCCUUUGGUCCUGCGGA .....((((((((((..(((((.(((((..(((((.((((....))))..)))))......((.......))(((((....))))).....))))).)))))))))))))))........ ( -40.10) >DroSim_CAF1 24222 120 - 1 UUGAUAUCAAGGGCAUCAGCUUCAUGUGGACCACUAUGGCCAUUGCCAGAAGUGGAGCGAUGCGGAUUAGGCGAUGUAAAUACAUCAUUAGCACAUUAAGUUUGCCUUUCGUCCUGCGGA ..(((...(((((((..(((((.(((((..(((((.((((....))))..)))))......((.......))(((((....))))).....))))).)))))))))))).)))....... ( -35.80) >DroYak_CAF1 20516 120 - 1 CGGAUAUCAGAGGCAUCAGCUUCAUGUGAACCACUGUGGCCAUCGCCAGGAGCGGAGCGAUGCGGAUGAGGCGGUGCAAAUACAUCAUUGGCACAUUAAGUUUGCCCUUGGUUCUGCGGG ((((.(((((.((((..(((((.(((((..(((.((((..(((((((....(((......)))......)))))))....))))....)))))))).))))))))).))))))))).... ( -37.40) >consensus UUGAUAUCAAGGGCAUCAGCUUCAUGUGGACCACUAUGGCCAUUGCCAGAAGUGGAGCGAUGCGGAUUAGGCGAUGUAAAUACAUCAUUAGCACAUUAAGUUUGCCUUUGGUCCUGCGGA .....((((((((((..(((((.(((((..(((((.((((....))))..)))))......((.......))((((......)))).....))))).)))))))))))))))........ (-35.88 = -36.25 + 0.38)

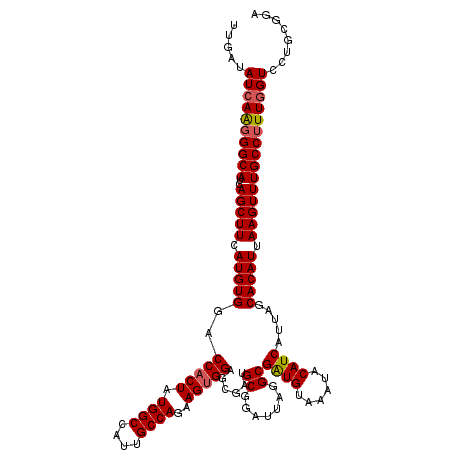
| Location | 8,640,409 – 8,640,529 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.44 |
| Mean single sequence MFE | -38.85 |
| Consensus MFE | -34.17 |
| Energy contribution | -33.11 |
| Covariance contribution | -1.06 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8640409 120 + 20766785 UUUACAUCGCUUAAUACGCAUCGCUCCACUUCUGGCAAUGGCCAUAGUGGUCCACAUGAAGCUGAUGCCCUUGAUAUCAAGUGGGCCACUCUUUGUAGGUGGUUAUAUAGGUAAUUCGGC ........(((((....(((((((((((((..((((....)))).))))).........))).)))))(((((....)))).)((((((.((....))))))))...)))))........ ( -33.30) >DroSec_CAF1 20585 120 + 1 UUUACAUCGCCUAAUCCGCAUCGCUCCACUUCUGGCAAUGGCCAUAGUGGUCCACAUGAAGCUGAUGCCCUUGAUAUCAAGUGGACCACUCUUUGUAGGUGGUUACAUAGGAAACGCGGC ...............((((......(((((((((((....)))).((((((((((.....((....))..(((....)))))))))))))....).)))))).......(....))))). ( -39.40) >DroSim_CAF1 24262 120 + 1 UUUACAUCGCCUAAUCCGCAUCGCUCCACUUCUGGCAAUGGCCAUAGUGGUCCACAUGAAGCUGAUGCCCUUGAUAUCAAGUGGACCACUCUUUGUAGGUGGUUACAUAGGAAACGCGGC ...............((((......(((((((((((....)))).((((((((((.....((....))..(((....)))))))))))))....).)))))).......(....))))). ( -39.40) >DroYak_CAF1 20556 120 + 1 UUUGCACCGCCUCAUCCGCAUCGCUCCGCUCCUGGCGAUGGCCACAGUGGUUCACAUGAAGCUGAUGCCUCUGAUAUCCGAUGGGCCACUCUUUGUGGGUGGGUAUCUCGGGAACGCGGC ......((((.(((...(((((((.(((((..((((....)))).)))))(((....))))).)))))...)))..(((((..(.((((((.....)))))).)...)))))...)))). ( -43.30) >consensus UUUACAUCGCCUAAUCCGCAUCGCUCCACUUCUGGCAAUGGCCAUAGUGGUCCACAUGAAGCUGAUGCCCUUGAUAUCAAGUGGACCACUCUUUGUAGGUGGUUACAUAGGAAACGCGGC ......((((....(((........((((((.((((....)))).(((((((((......((....)).((((....)))))))))))))......)))))).......)))...)))). (-34.17 = -33.11 + -1.06)



| Location | 8,640,409 – 8,640,529 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.44 |
| Mean single sequence MFE | -39.40 |
| Consensus MFE | -33.17 |
| Energy contribution | -32.67 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8640409 120 - 20766785 GCCGAAUUACCUAUAUAACCACCUACAAAGAGUGGCCCACUUGAUAUCAAGGGCAUCAGCUUCAUGUGGACCACUAUGGCCAUUGCCAGAAGUGGAGCGAUGCGUAUUAAGCGAUGUAAA ......................(((((..((((.((((..(((....)))))))....))))..))))).(((((.((((....))))..))))).(((.(((.......))).)))... ( -33.40) >DroSec_CAF1 20585 120 - 1 GCCGCGUUUCCUAUGUAACCACCUACAAAGAGUGGUCCACUUGAUAUCAAGGGCAUCAGCUUCAUGUGGACCACUAUGGCCAUUGCCAGAAGUGGAGCGAUGCGGAUUAGGCGAUGUAAA ...(((((.((((.....((((((....)).))))(((.((((....)))).(((((.((((((((((...)))..((((....))))...))))))))))))))).)))).)))))... ( -41.80) >DroSim_CAF1 24262 120 - 1 GCCGCGUUUCCUAUGUAACCACCUACAAAGAGUGGUCCACUUGAUAUCAAGGGCAUCAGCUUCAUGUGGACCACUAUGGCCAUUGCCAGAAGUGGAGCGAUGCGGAUUAGGCGAUGUAAA ...(((((.((((.....((((((....)).))))(((.((((....)))).(((((.((((((((((...)))..((((....))))...))))))))))))))).)))).)))))... ( -41.80) >DroYak_CAF1 20556 120 - 1 GCCGCGUUCCCGAGAUACCCACCCACAAAGAGUGGCCCAUCGGAUAUCAGAGGCAUCAGCUUCAUGUGAACCACUGUGGCCAUCGCCAGGAGCGGAGCGAUGCGGAUGAGGCGGUGCAAA (((((....((((.....((((.(.....).))))....))))...(((...(((((.(((((..(((...)))(.((((....)))).)...))))))))))...))).)))))..... ( -40.60) >consensus GCCGCGUUUCCUAUAUAACCACCUACAAAGAGUGGCCCACUUGAUAUCAAGGGCAUCAGCUUCAUGUGGACCACUAUGGCCAUUGCCAGAAGUGGAGCGAUGCGGAUUAGGCGAUGUAAA (((....(((........(((((......).))))....((((....)))).(((((.((((((((((...)))..((((....))))...)))))))))))))))...)))........ (-33.17 = -32.67 + -0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:12 2006