
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,639,715 – 8,639,912 |
| Length | 197 |
| Max. P | 0.971281 |

| Location | 8,639,715 – 8,639,832 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.48 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -25.27 |
| Energy contribution | -26.15 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971281 |
| Prediction | RNA |
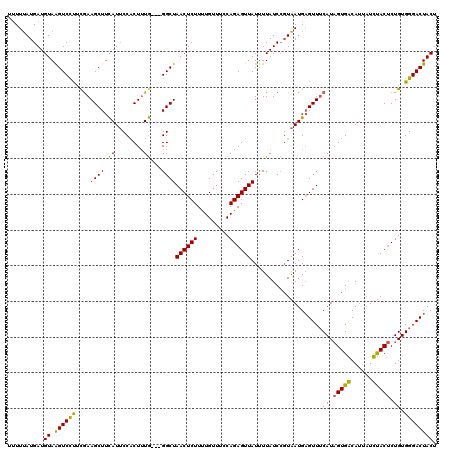
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8639715 117 - 20766785 UAAGGAUAAAAUAACUCUGGAAACAAAAGAGUUAGCC---CAAAGUGGAAUGAAGCUUCGAAGGACUUACAUCAUAAAAAUGGUGAGGGCAUCCCAGGGCUCACUAUCACUGGUCUGACA ...((((....((((((((....)...)))))))(((---(...........(((..((....))))).((((((....)))))).))))))))((((.(...........).))))... ( -30.00) >DroSec_CAF1 19891 117 - 1 UACGGAUAAAAUAACUCUGGAAACAAAAGAGUUAGCC---CAAAGUGGAAUGAAGCUUUAAAGGACCUACAUCAUAAAUAUGGUGAGUCCAUCCCAGGGCUCACUAUCACUGGUCUGGCA ...........((((((((....)...)))))))(((---.(((((........)))))...(((((............(((((((((((......)))))))))))....)))))))). ( -35.49) >DroSim_CAF1 23568 117 - 1 UACGGAUAAAAUAACUCUGGAAACAAAAGAGUUAGCC---CACAGUGGAAUGAAGCUUCAAAGGACCUACAUCAUAAAAAUGGUGAGUCCAUCCCAGGGCUCACUAUCACUGGUCUGGCA ..(((((....((((((((....)...)))))))...---..(((((((.((.((.((.....)).)).))))......(((((((((((......)))))))))))))))))))))... ( -37.00) >DroYak_CAF1 19859 120 - 1 CACCGAUAACAUAACUCUGCAAACAGAAGAGUUAGCCAUCUAAAGCGGUAUAAAGCUUCGGGAGACUUACAUCAUGAAAAUGGUGAGUCCAUCCCAGGACCCACUUUCACUCGUCUGGCA ...........(((((((.........)))))))(((.((..((((........))))..))((((...((((((....)))))).((((......))))............))))))). ( -34.80) >consensus UACGGAUAAAAUAACUCUGGAAACAAAAGAGUUAGCC___CAAAGUGGAAUGAAGCUUCAAAGGACCUACAUCAUAAAAAUGGUGAGUCCAUCCCAGGGCUCACUAUCACUGGUCUGGCA ...........((((((((....)...)))))))(((.....((((........))))....(((((.....(((....)))((((((((......)))))))).......)))))))). (-25.27 = -26.15 + 0.88)

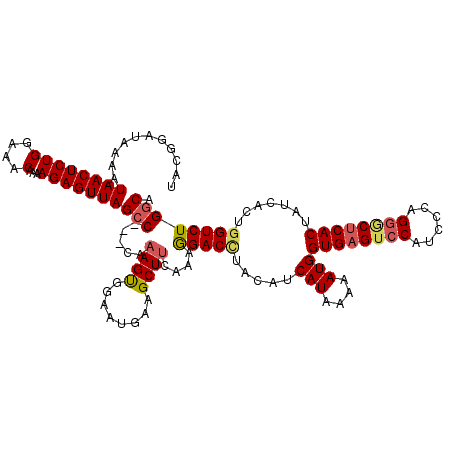

| Location | 8,639,755 – 8,639,872 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.78 |
| Mean single sequence MFE | -28.06 |
| Consensus MFE | -18.31 |
| Energy contribution | -16.88 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8639755 117 + 20766785 UUUUUAUGAUGUAAGUCCUUCGAAGCUUCAUUCCACUUUG---GGCUAACUCUUUUGUUUCCAGAGUUAUUUUAUCCUUAAUGAGUUUCAUAGUGGCAUUAUCUACUCUGUGGGACUACU ..........(((.(((((.(((((((.((((..((((((---((..(((......)))))))))))............))))))))))..(((((......)))))..).)))))))). ( -26.24) >DroSec_CAF1 19931 117 + 1 UAUUUAUGAUGUAGGUCCUUUAAAGCUUCAUUCCACUUUG---GGCUAACUCUUUUGUUUCCAGAGUUAUUUUAUCCGUAAUGAGUUUCAUAGUGACAUUAUUUACUCUGUGGGACUACU ...((((((((.((((........)))))))...((((((---((..(((......))))))))))).........)))))..(((..(((((.(..........).)))))..)))... ( -22.50) >DroSim_CAF1 23608 117 + 1 UUUUUAUGAUGUAGGUCCUUUGAAGCUUCAUUCCACUGUG---GGCUAACUCUUUUGUUUCCAGAGUUAUUUUAUCCGUAAUGUGUUUCAUAGUGACAUUAUUUACUCUGUGGGACUACU ..........(((((((((.((((((..((((.(...(((---((.(((((((.........))))))).)))))..).)))).)))))).)).)))........((.....)).)))). ( -23.90) >DroYak_CAF1 19899 120 + 1 UUUUCAUGAUGUAAGUCUCCCGAAGCUUUAUACCGCUUUAGAUGGCUAACUCUUCUGUUUGCAGAGUUAUGUUAUCGGUGAUGAGUUGCAUUGUGGCAUUCUCCACCCUGUGGGACUACU ...(((..((((((.(((((((((((........))))).(((((((((((((.(.....).))))))).)))))))).)).)).))))))..)))...((.((((...))))))..... ( -39.60) >consensus UUUUUAUGAUGUAAGUCCUUCGAAGCUUCAUUCCACUUUG___GGCUAACUCUUUUGUUUCCAGAGUUAUUUUAUCCGUAAUGAGUUUCAUAGUGACAUUAUCUACUCUGUGGGACUACU ..........((.((((((....((((.((........))...))))((((((.........))))))....................((.(((((......))))).)).)))))))). (-18.31 = -16.88 + -1.44)

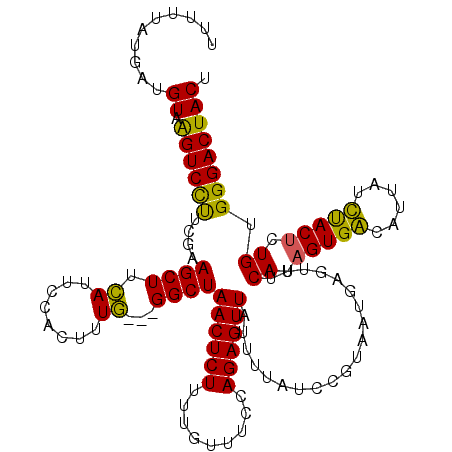
| Location | 8,639,755 – 8,639,872 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.78 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -17.48 |
| Energy contribution | -17.79 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8639755 117 - 20766785 AGUAGUCCCACAGAGUAGAUAAUGCCACUAUGAAACUCAUUAAGGAUAAAAUAACUCUGGAAACAAAAGAGUUAGCC---CAAAGUGGAAUGAAGCUUCGAAGGACUUACAUCAUAAAAA .(((((((..(..(((........((((((((.....)))...((......((((((((....)...)))))))..)---)..)))))......)))..)..)))).))).......... ( -25.34) >DroSec_CAF1 19931 117 - 1 AGUAGUCCCACAGAGUAAAUAAUGUCACUAUGAAACUCAUUACGGAUAAAAUAACUCUGGAAACAAAAGAGUUAGCC---CAAAGUGGAAUGAAGCUUUAAAGGACCUACAUCAUAAAUA .((((..((.((((((......((((...(((.....)))....)))).....))))))......((((..(((..(---(.....))..)))..))))...))..)))).......... ( -25.30) >DroSim_CAF1 23608 117 - 1 AGUAGUCCCACAGAGUAAAUAAUGUCACUAUGAAACACAUUACGGAUAAAAUAACUCUGGAAACAAAAGAGUUAGCC---CACAGUGGAAUGAAGCUUCAAAGGACCUACAUCAUAAAAA .((((..((.((((((......((((...(((.....)))....)))).....)))))).......(((..(((..(---(.....))..)))..)))....))..)))).......... ( -22.80) >DroYak_CAF1 19899 120 - 1 AGUAGUCCCACAGGGUGGAGAAUGCCACAAUGCAACUCAUCACCGAUAACAUAACUCUGCAAACAGAAGAGUUAGCCAUCUAAAGCGGUAUAAAGCUUCGGGAGACUUACAUCAUGAAAA .((((((((....((((..((.(((......)))..))..))))(((....(((((((.........)))))))...)))..((((........)))).))))...)))).......... ( -28.10) >consensus AGUAGUCCCACAGAGUAAAUAAUGCCACUAUGAAACUCAUUACGGAUAAAAUAACUCUGGAAACAAAAGAGUUAGCC___CAAAGUGGAAUGAAGCUUCAAAGGACCUACAUCAUAAAAA .(((((((....((((........((((((((.....)))...........((((((((....)...))))))).........)))))......))))....)))).))).......... (-17.48 = -17.79 + 0.31)



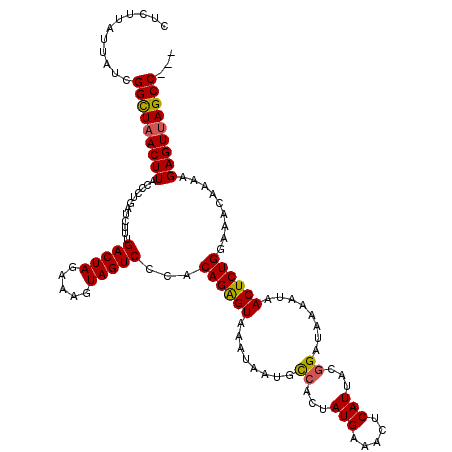
| Location | 8,639,795 – 8,639,912 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.19 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -22.85 |
| Energy contribution | -22.66 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8639795 117 - 20766785 CUCUUAUUAUCGGUUAACUUACCCUGAUCUUUGACUAGAAAGUAGUCCCACAGAGUAGAUAAUGCCACUAUGAAACUCAUUAAGGAUAAAAUAACUCUGGAAACAAAAGAGUUAGCC--- ...........(((((((((............(((((.....)))))...((((((........((...(((.....)))...))........)))))).........)))))))))--- ( -24.89) >DroSec_CAF1 19971 117 - 1 CUCUUAUUAGCGGCUAACUUACCCUGAUCUUUGACUAGAAAGUAGUCCCACAGAGUAAAUAAUGUCACUAUGAAACUCAUUACGGAUAAAAUAACUCUGGAAACAAAAGAGUUAGCC--- ...........(((((((((............(((((.....)))))...((((((......((((...(((.....)))....)))).....)))))).........)))))))))--- ( -27.30) >DroSim_CAF1 23648 117 - 1 CUCUUAUUAUCGGCUAACUUACCCUGAUCUUUGACUAGAAAGUAGUCCCACAGAGUAAAUAAUGUCACUAUGAAACACAUUACGGAUAAAAUAACUCUGGAAACAAAAGAGUUAGCC--- ...........(((((((((............(((((.....)))))...((((((......((((...(((.....)))....)))).....)))))).........)))))))))--- ( -26.30) >DroYak_CAF1 19939 120 - 1 GGUUAAUAAUCGGUUCACUUACUCUGAUCUUUGACUAGGAAGUAGUCCCACAGGGUGGAGAAUGCCACAAUGCAACUCAUCACCGAUAACAUAACUCUGCAAACAGAAGAGUUAGCCAUC (((((((.((((((.....(((((((......(((((.....)))))...)))))))(((..(((......))).)))...))))))........((((....))))...)))))))... ( -31.40) >consensus CUCUUAUUAUCGGCUAACUUACCCUGAUCUUUGACUAGAAAGUAGUCCCACAGAGUAAAUAAUGCCACUAUGAAACUCAUUACGGAUAAAAUAACUCUGGAAACAAAAGAGUUAGCC___ ...........(((((((((............(((((.....)))))...((((((........((...(((.....)))...))........)))))).........)))))))))... (-22.85 = -22.66 + -0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:08 2006