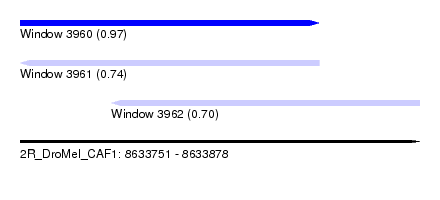
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,633,751 – 8,633,878 |
| Length | 127 |
| Max. P | 0.965549 |

| Location | 8,633,751 – 8,633,846 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 88.85 |
| Mean single sequence MFE | -25.47 |
| Consensus MFE | -22.17 |
| Energy contribution | -22.40 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
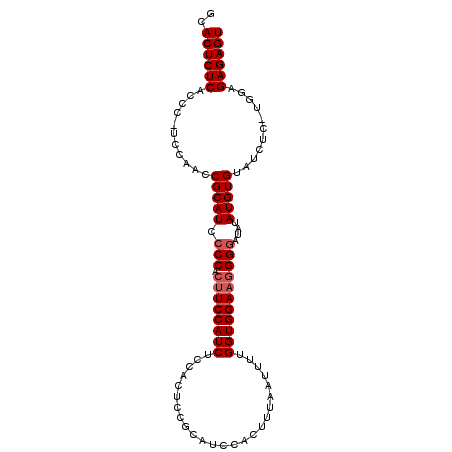
>2R_DroMel_CAF1 8633751 95 + 20766785 CCUCUCUCGCCAUGCUCCAUCCCACUCGCACUCUCUCCA-GAGAUACACAUAUAUCCCUUUCCACCAAAAUUUAAGUGGAUGCGGCGUGGAGAUGG ................((((((((((((((.((((....-))))................(((((..........)))))))))).)))).))))) ( -27.50) >DroSec_CAF1 13911 96 + 1 CCCCUCUCGCCAUACUCCAUCCCACUCGCACUCUCUUCAGGACAUACACAUAUAUUCCCUUCCACCAAAAUUAAUGUGGAUGCGGAGUGGAGAUGG ................((((((((((((((.........(((.(((......))))))..(((((..........)))))))).)))))).))))) ( -25.90) >DroSim_CAF1 17528 95 + 1 CCCCUCUCGGCAUGCUCUAUCCCACUUACACUCUCUCUA-GAGAUACACAUACAUCCCCUUCCACCAAAAUUAAAGUGGAUGCGGAGUGGAGAUGG ................(((((((((......((((....-))))..........(((.(.(((((..........))))).).))))))).))))) ( -23.00) >consensus CCCCUCUCGCCAUGCUCCAUCCCACUCGCACUCUCUCCA_GAGAUACACAUAUAUCCCCUUCCACCAAAAUUAAAGUGGAUGCGGAGUGGAGAUGG ................((((((((((((((.((((.....))))................(((((..........)))))))))).)))).))))) (-22.17 = -22.40 + 0.23)


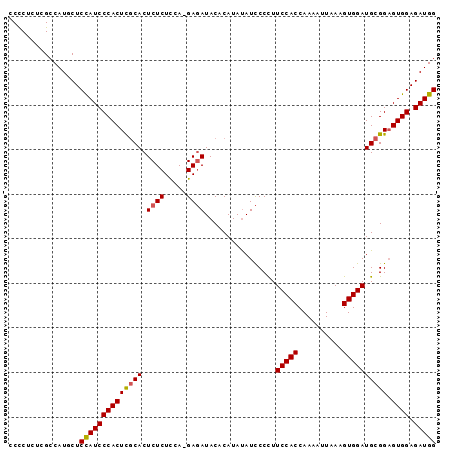
| Location | 8,633,751 – 8,633,846 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 88.85 |
| Mean single sequence MFE | -33.16 |
| Consensus MFE | -25.22 |
| Energy contribution | -26.33 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740712 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8633751 95 - 20766785 CCAUCUCCACGCCGCAUCCACUUAAAUUUUGGUGGAAAGGGAUAUAUGUGUAUCUC-UGGAGAGAGUGCGAGUGGGAUGGAGCAUGGCGAGAGAGG (((((.((((..((((((((((........)))))).((((((((....)))))))-)........)))).)))))))))................ ( -35.30) >DroSec_CAF1 13911 96 - 1 CCAUCUCCACUCCGCAUCCACAUUAAUUUUGGUGGAAGGGAAUAUAUGUGUAUGUCCUGAAGAGAGUGCGAGUGGGAUGGAGUAUGGCGAGAGGGG (((((.(((((((((.(((((..........)))))(((..((((....))))..))).......))).))))))))))).......(....)... ( -32.40) >DroSim_CAF1 17528 95 - 1 CCAUCUCCACUCCGCAUCCACUUUAAUUUUGGUGGAAGGGGAUGUAUGUGUAUCUC-UAGAGAGAGUGUAAGUGGGAUAGAGCAUGCCGAGAGGGG ((.((((...(((.(.((((((........)))))).).))).((((((.((((((-................))))))..)))))).)))).)). ( -31.79) >consensus CCAUCUCCACUCCGCAUCCACUUUAAUUUUGGUGGAAGGGGAUAUAUGUGUAUCUC_UGGAGAGAGUGCGAGUGGGAUGGAGCAUGGCGAGAGGGG (((((((((..((((.((((((........))))))..(((((((....)))))))...............))))..))))).))))(....)... (-25.22 = -26.33 + 1.11)

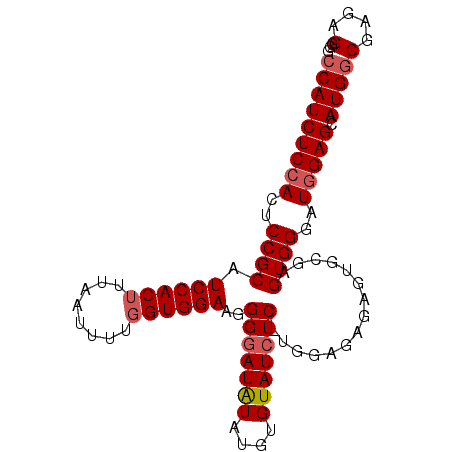
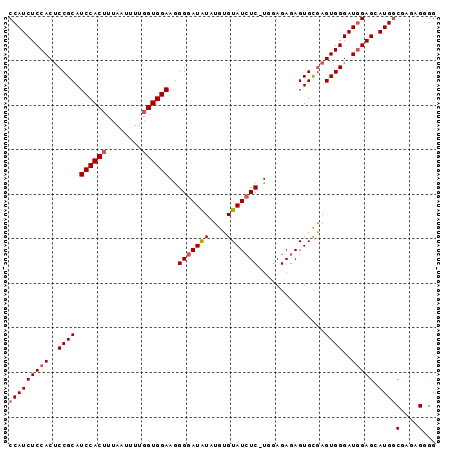
| Location | 8,633,780 – 8,633,878 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 90.51 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -19.86 |
| Energy contribution | -20.86 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8633780 98 - 20766785 GCACUCUCACCCCCCCAACCGCAUCCCCACUUCCAUCUCCACGCCGCAUCCACUUAAAUUUUGGUGGAAAGGGAUAUAUGUGUAUCUC-UGGAGAGAGU ..((((((.((.....................................((((((........)))))).((((((((....)))))))-))).)))))) ( -25.30) >DroSec_CAF1 13940 98 - 1 GCACUCUCACCC-UCCACCCGCAUCCCCACUUCCAUCUCCACUCCGCAUCCACAUUAAUUUUGGUGGAAGGGAAUAUAUGUGUAUGUCCUGAAGAGAGU ..((((((....-..((..(((((..((.((((((((.........................)))))))))).....)))))..)).......)))))) ( -22.93) >DroSim_CAF1 17557 97 - 1 GCACUCUCACCC-UCCAACCGCAUCCCCACCUCCAUCUCCACUCCGCAUCCACUUUAAUUUUGGUGGAAGGGGAUGUAUGUGUAUCUC-UAGAGAGAGU ..((((((....-..((...((((((((...((((((.........................)))))).))))))))...))..((..-..)))))))) ( -27.91) >consensus GCACUCUCACCC_UCCAACCGCAUCCCCACUUCCAUCUCCACUCCGCAUCCACUUUAAUUUUGGUGGAAGGGGAUAUAUGUGUAUCUC_UGGAGAGAGU ..((((((...........(((((.(((.((((((((.........................)))))))))))....)))))...........)))))) (-19.86 = -20.86 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:59 2006