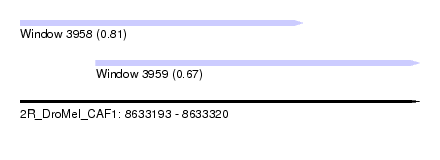
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,633,193 – 8,633,320 |
| Length | 127 |
| Max. P | 0.807875 |

| Location | 8,633,193 – 8,633,283 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 86.40 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -19.30 |
| Energy contribution | -20.05 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
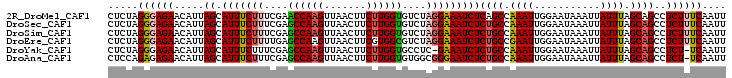
>2R_DroMel_CAF1 8633193 90 + 20766785 CUCCCUCUUUAAAA--------GCACUUCUCCCUCUAGGGAGAACAUUAGCAUUUCUUUCGAACCAAGUUAACUUCUUGGUGUCUAGGAAAUCUCAGC ..............--------((..(((((((....)))))))....((.(((((((....((((((.......))))))....)))))))))..)) ( -24.30) >DroSec_CAF1 13352 90 + 1 CUCCCUCUUUAAAA--------GCACUUCUCCCUCUAGGGAGAACAUUAGCAUUUCUUUCGAGCCAAGUUAACUUCUUGGUGUCUAGGAAAUCUCUGC ..............--------(((.(((((((....)))))))....((.(((((((....((((((.......))))))....))))))))).))) ( -24.90) >DroSim_CAF1 16969 90 + 1 CUCCCUCUUUAAAA--------GCACAUCUCCCUCUAGGGAGAACAUUAGCAUUUCUUUCGAGCCAAGUUAACUUCUUGGUGUCUAGGAAAUCUCUGC ..............--------(((..((((((....)))))).....((.(((((((....((((((.......))))))....))))))))).))) ( -24.50) >DroEre_CAF1 13290 90 + 1 UUCCCUCUUUAAAG--------ACGCUUCUCCCUCUAGGGAGAACAUUAGCAUUUCUUUUGAGCCAAGUUAACUUCGUGGCGUCUAGGAAAUCUCUGC ...(((......((--------(((((((((((....)))))).....)))...........((((.(.......).))))))))))).......... ( -22.80) >DroYak_CAF1 13330 89 + 1 UUCCCUCUUUAAAA--------GCACUUCUCCCUCUAGGGAGAACAUUAGCAUUUCUUUCGAGCCAAGUUAACUUCUUGGUGCCUC-GAAAUCUCUGC ..............--------....(((((((....))))))).....(((....((((((((((((.......)))))...)))-))))....))) ( -24.50) >DroAna_CAF1 14207 98 + 1 GGUCGUCUUCAGAGAAUCCCUCGUGCCGCUACCUCCAGAGAGAACAUUAGCAUUUCUUUCGAGCCAAGUUAACUUCUUGGUGUGGCGGGAAUCUCUGC .........((((((.((((....(((((........(((((((.........)))))))..((((((.......))))))))))))))).)))))). ( -34.90) >consensus CUCCCUCUUUAAAA________GCACUUCUCCCUCUAGGGAGAACAUUAGCAUUUCUUUCGAGCCAAGUUAACUUCUUGGUGUCUAGGAAAUCUCUGC ...........................((((((....)))))).....((.(((((((....((((((.......))))))....))))))))).... (-19.30 = -20.05 + 0.75)

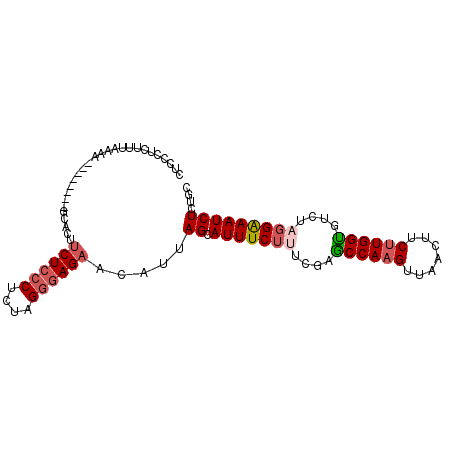
| Location | 8,633,217 – 8,633,320 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -22.23 |
| Consensus MFE | -20.04 |
| Energy contribution | -20.52 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8633217 103 + 20766785 CUCUAGGGAGAACAUUAGCAUUUCUUUCGAACCAAGUUAACUUCUUGGUGUCUAGGAAAUCUCAGCCAAAUUGGAAUAAAUUAUUUAGCAGCCUCUUUCAAUU .....(..(((.....((.(((((((....((((((.......))))))....)))))))))..((.((((...........)))).))....)))..).... ( -19.80) >DroSec_CAF1 13376 103 + 1 CUCUAGGGAGAACAUUAGCAUUUCUUUCGAGCCAAGUUAACUUCUUGGUGUCUAGGAAAUCUCUGCCAAAUUGGAAUAAAUUAUUUAGCAGCCUCUUUCAAUU .....(..(((.....((.(((((((....((((((.......))))))....)))))))))((((.((((...........)))).))))..)))..).... ( -22.90) >DroSim_CAF1 16993 103 + 1 CUCUAGGGAGAACAUUAGCAUUUCUUUCGAGCCAAGUUAACUUCUUGGUGUCUAGGAAAUCUCUGCCAAAUUGGAAUAAAUUAUUUAGCAGCCUCUUUCAAUU .....(..(((.....((.(((((((....((((((.......))))))....)))))))))((((.((((...........)))).))))..)))..).... ( -22.90) >DroEre_CAF1 13314 103 + 1 CUCUAGGGAGAACAUUAGCAUUUCUUUUGAGCCAAGUUAACUUCGUGGCGUCUAGGAAAUCUCUGCCGAAUUGGAAUAAAUUAUUUAGCAGCCUCUUUCAAUU .....(..(((.....((.(((((((....((((.(.......).))))....)))))))))((((..(((...........)))..))))..)))..).... ( -21.50) >DroYak_CAF1 13354 101 + 1 CUCUAGGGAGAACAUUAGCAUUUCUUUCGAGCCAAGUUAACUUCUUGGUGCCUC-GAAAUCUCUGCCAAAUUGGAAUAAAUUAUUUAGCAGCCUCU-UCAAUU ......(((((.............((((((((((((.......)))))...)))-))))...((((.((((...........)))).))))..)))-)).... ( -21.00) >DroAna_CAF1 14239 102 + 1 CUCCAGAGAGAACAUUAGCAUUUCUUUCGAGCCAAGUUAACUUCUUGGUGUGGCGGGAAUCUCUGCCAAAUUGGAAUAAAUUAUUUAGCAGCCUCU-UCAAUU .(((((((((((.........)))))))..((((((.......)))))).(((((((....)))))))...)))).....................-...... ( -25.30) >consensus CUCUAGGGAGAACAUUAGCAUUUCUUUCGAGCCAAGUUAACUUCUUGGUGUCUAGGAAAUCUCUGCCAAAUUGGAAUAAAUUAUUUAGCAGCCUCUUUCAAUU .....((((((.....((.(((((((....((((((.......))))))....)))))))))((((.((((...........)))).))))..)))))).... (-20.04 = -20.52 + 0.48)
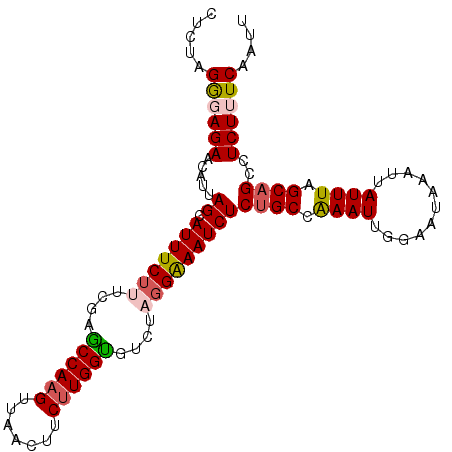
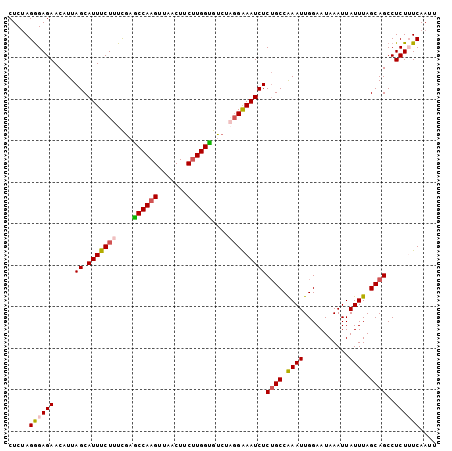
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:57 2006