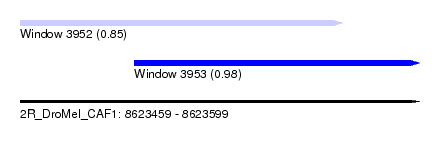
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,623,459 – 8,623,599 |
| Length | 140 |
| Max. P | 0.975902 |

| Location | 8,623,459 – 8,623,572 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 86.45 |
| Mean single sequence MFE | -38.45 |
| Consensus MFE | -30.14 |
| Energy contribution | -30.00 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
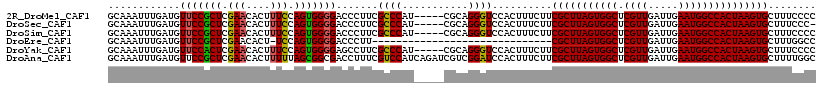
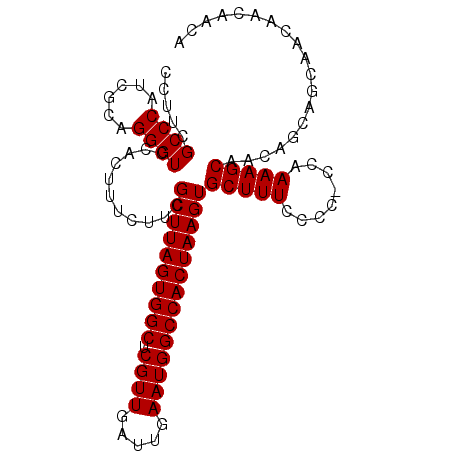
>2R_DroMel_CAF1 8623459 113 + 20766785 GCAAAUUUGAUGUUCCGCUCGAACACUUUCCAGUGGGGACCCUUCGCCCAU-----CGCAGGGUCCACUUUCUUCGCUUAGUGGCUCGUUGAUUGAAUGGCCACUAAGUGCUUUCCCC ((......((....(((((.(((....))).)))))(((((((.((.....-----)).))))))).......))((((((((((.((((.....))))))))))))))))....... ( -42.20) >DroSec_CAF1 3135 112 + 1 GCAAAUUUGAUGUUCCGCUCGAACACUUUCCAGUGGGGACCCUUCGCCCAU-----CGCAGGGUCCACUUUCUUCGCUUAGUGGCUCGUUGAUUGAAUGGCCACUAAGUGCUUUCCC- ((......((....(((((.(((....))).)))))(((((((.((.....-----)).))))))).......))((((((((((.((((.....))))))))))))))))......- ( -42.20) >DroSim_CAF1 3074 113 + 1 GCAAAUUUGAUGUUCCGCUCGAACACUUUCCAGUGGGGACCCUUCGCCCAU-----CGCAGGGUCCACUUUCUUCGCUUAGUGGCUCGUUGAUUGAAUGGCCACUAAGUGCUUUCCCC ((......((....(((((.(((....))).)))))(((((((.((.....-----)).))))))).......))((((((((((.((((.....))))))))))))))))....... ( -42.20) >DroEre_CAF1 3739 87 + 1 GCAAAUUUGAUGUUCCGCUCGAACACU-UCCAGUGGGGACCCUU------------------------------CGCUUAGUGGCUCGUUGAUUGAAUGGCCACUAAGUGCUUUGGCC ..............(((((.((.....-)).)))))((.((...------------------------------(((((((((((.((((.....)))))))))))))))....)))) ( -29.80) >DroYak_CAF1 3512 113 + 1 GCAAAUUUGAUGUUCCACUCGAACACUUUCCAGUGGGGAGCCUUCGCCCAU-----CGCAGGGUCCACUUUCUUCGCUUAGUGGCUCGUUGAUUGAAUGGCCACUAAGUGCUUUCCCC ((......((....(((((.(((....))).)))))(((.(((.((.....-----)).))).))).......))((((((((((.((((.....))))))))))))))))....... ( -38.10) >DroAna_CAF1 3305 118 + 1 GCAAAUUUGAUGUUCCGCUCGAACACUUUUUAGCGGCGACCUUUCGUCCAUCAGAUCGUCGGAUCCACUUUCUUCGCUUAGUGGCUCGUUGAUUGAAUGGCCACUAAGUGCUUUUGGC ....((((((((..(((((.(((....))).))))).(((.....))))))))))).((((((......)))..(((((((((((.((((.....))))))))))))))).....))) ( -36.20) >consensus GCAAAUUUGAUGUUCCGCUCGAACACUUUCCAGUGGGGACCCUUCGCCCAU_____CGCAGGGUCCACUUUCUUCGCUUAGUGGCUCGUUGAUUGAAUGGCCACUAAGUGCUUUCCCC ............(((((((.(((....))).))))))).......((((...........))))..........(((((((((((.((((.....)))))))))))))))........ (-30.14 = -30.00 + -0.14)
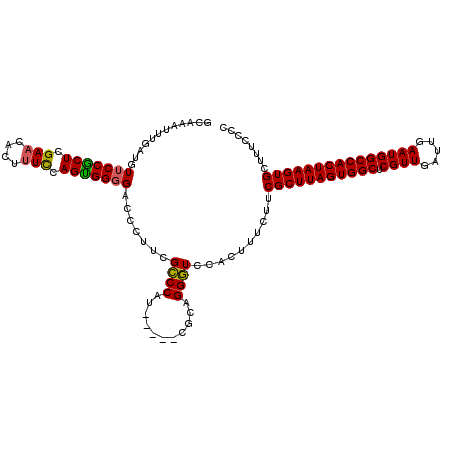
| Location | 8,623,499 – 8,623,599 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 96.35 |
| Mean single sequence MFE | -28.03 |
| Consensus MFE | -27.40 |
| Energy contribution | -27.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8623499 100 + 20766785 CCUUCGCCCAUCGCAGGGUCCACUUUCUUCGCUUAGUGGCUCGUUGAUUGAAUGGCCACUAAGUGCUUUCCCC-CCAAAAGCAACAGCAACAACAACAAGA .....((((......))))......((((.((((((((((.((((.....))))))))))))))(((((....-...)))))...............)))) ( -27.50) >DroSec_CAF1 3175 99 + 1 CCUUCGCCCAUCGCAGGGUCCACUUUCUUCGCUUAGUGGCUCGUUGAUUGAAUGGCCACUAAGUGCUUUCCC--CCAAAAGCAGCAGCAGCAACAACAACA .....((((......))))...........((((((((((.((((.....))))))))))))))(((((...--...))))).((....)).......... ( -28.80) >DroSim_CAF1 3114 101 + 1 CCUUCGCCCAUCGCAGGGUCCACUUUCUUCGCUUAGUGGCUCGUUGAUUGAAUGGCCACUAAGUGCUUUCCCCGCCAAAAGCAGCAGCAGCAACAACAACA .....((((......))))...........((((((((((.((((.....))))))))))))))(((((........))))).((....)).......... ( -28.60) >DroYak_CAF1 3552 100 + 1 CCUUCGCCCAUCGCAGGGUCCACUUUCUUCGCUUAGUGGCUCGUUGAUUGAAUGGCCACUAAGUGCUUUCCCC-CCAAAAGCAACAAAAGCAACAACAACA .....((((......))))...((((....((((((((((.((((.....))))))))))))))(((((....-...)))))....))))........... ( -27.20) >consensus CCUUCGCCCAUCGCAGGGUCCACUUUCUUCGCUUAGUGGCUCGUUGAUUGAAUGGCCACUAAGUGCUUUCCCC_CCAAAAGCAACAGCAGCAACAACAACA .....((((......))))...........((((((((((.((((.....))))))))))))))(((((........)))))................... (-27.40 = -27.40 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:51 2006