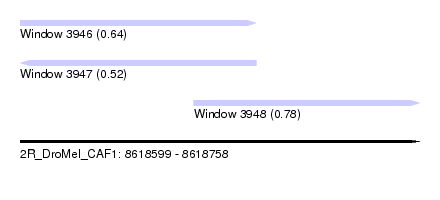
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,618,599 – 8,618,758 |
| Length | 159 |
| Max. P | 0.782958 |

| Location | 8,618,599 – 8,618,693 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.49 |
| Mean single sequence MFE | -32.84 |
| Consensus MFE | -13.78 |
| Energy contribution | -14.58 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.641524 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8618599 94 + 20766785 AAUUGCAAUUGGAGGACUCUGCAGAUGCAAUUGGCAGUGCAUCGUAGGA---C----CCUCCAAUGAAGUU----UCCGAAACUGAGUGUCCUAUAUACGACAGA--------------- ((((...((((((((..(((((.((((((........))))))))))).---.----))))))))..))))----.......(((.((((.....))))..))).--------------- ( -29.60) >DroSec_CAF1 20890 107 + 1 AAUUGUAAUUGGAGGACUCUGCAGAUGCAA-UGGCAGUGCAUCGGAGGA---C----CCUCCAAUGAAGUU----UCCGAAAGUGAGUGUCCUAUAUACGAAGAAUCUCGCUCACUGAG- ((((...((((((((..(((.(.((((((.-......))))))).))).---.----))))))))..))))----......((((((((...................))))))))...- ( -32.81) >DroSim_CAF1 18180 107 + 1 AAUUGUAAUUGGAGGACUCUGCAGAUGGAA-UGGCAGUGCAUCGGAGGA---C----CCUCCAAUGAAGUU----UGCGAAACUGAGUGUCCUAUAUACGACGAUUCUCGCACACUGAG- .......((((((((.(((((((..((...-...)).)))...))))..---.----))))))))..(((.----(((((....(((((((........))).))))))))).)))...- ( -30.20) >DroEre_CAF1 18634 115 + 1 AACUGUAAUUGGAAGUCCCUGCAGCUGGCUGUCCUUGUCAGUUGGAGGAGAGCCCUGCCAUCAUUGGAGUGCUGCACCAGCACUCAAUUUUUGAUAAACGAAA---CCCG--CACCGGGA .......((..(((((....((((..((((.(((((........))))).))))))))........((((((((...)))))))).)))))..))........---((((--...)))). ( -39.90) >DroYak_CAF1 14396 104 + 1 AACUGUAAUUCGAGGACUCUGCAGAUGCAACUGCCAGUGCAUCGGAGGA---C----CCUUCAAUGAAGUU----UCCGAAACUGAGUGUCCUAUAUACGAAG---CUCG--CACUCAGU ........((((..((((...((((((((........))))))((((..---.----.))))..)).))))----..))))(((((((((.((........))---...)--)))))))) ( -31.70) >consensus AAUUGUAAUUGGAGGACUCUGCAGAUGCAA_UGGCAGUGCAUCGGAGGA___C____CCUCCAAUGAAGUU____UCCGAAACUGAGUGUCCUAUAUACGAAG___CUCG__CACUGAG_ ............(((((.((...((((((........))))))(((((.........))))).......................)).)))))........................... (-13.78 = -14.58 + 0.80)

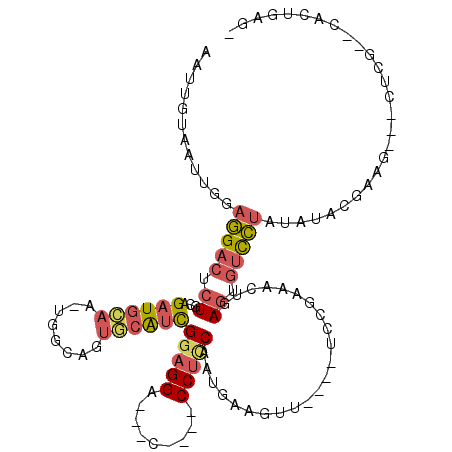
| Location | 8,618,599 – 8,618,693 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.49 |
| Mean single sequence MFE | -31.12 |
| Consensus MFE | -8.05 |
| Energy contribution | -9.77 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.26 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.515875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8618599 94 - 20766785 ---------------UCUGUCGUAUAUAGGACACUCAGUUUCGGA----AACUUCAUUGGAGG----G---UCCUACGAUGCACUGCCAAUUGCAUCUGCAGAGUCCUCCAAUUGCAAUU ---------------..((((........)))).........(..----..)...((((((((----(---..((.(((((((........)))))).).))..)))))))))....... ( -26.60) >DroSec_CAF1 20890 107 - 1 -CUCAGUGAGCGAGAUUCUUCGUAUAUAGGACACUCACUUUCGGA----AACUUCAUUGGAGG----G---UCCUCCGAUGCACUGCCA-UUGCAUCUGCAGAGUCCUCCAAUUACAAUU -...((((((.(...((((........))))).))))))...(..----..)...((((((((----(---..(((.((((((......-.)))))).).))..)))))))))....... ( -31.80) >DroSim_CAF1 18180 107 - 1 -CUCAGUGUGCGAGAAUCGUCGUAUAUAGGACACUCAGUUUCGCA----AACUUCAUUGGAGG----G---UCCUCCGAUGCACUGCCA-UUCCAUCUGCAGAGUCCUCCAAUUACAAUU -....((((((((......))))))))(((((.(((((....(((----.....((((((((.----.---..))))))))...)))..-......))).)).)))))............ ( -30.20) >DroEre_CAF1 18634 115 - 1 UCCCGGUG--CGGG---UUUCGUUUAUCAAAAAUUGAGUGCUGGUGCAGCACUCCAAUGAUGGCAGGGCUCUCCUCCAACUGACAAGGACAGCCAGCUGCAGGGACUUCCAAUUACAGUU ......((--..((---(..(...(((((......((((((((...))))))))...)))))((((((((.((((..........)))).))))..)))).)..)))..))......... ( -35.90) >DroYak_CAF1 14396 104 - 1 ACUGAGUG--CGAG---CUUCGUAUAUAGGACACUCAGUUUCGGA----AACUUCAUUGAAGG----G---UCCUCCGAUGCACUGGCAGUUGCAUCUGCAGAGUCCUCGAAUUACAGUU (((((((.--((((---.(((((((..(((((..(((((...(..----..)...)))))...----)---))))...))))....((((......)))).)))..)))).))).)))). ( -31.10) >consensus _CUCAGUG__CGAG___CUUCGUAUAUAGGACACUCAGUUUCGGA____AACUUCAUUGGAGG____G___UCCUCCGAUGCACUGCCA_UUGCAUCUGCAGAGUCCUCCAAUUACAAUU .....................(((...(((((..........(((........((......))........)))...((((((........))))))......))))).....))).... ( -8.05 = -9.77 + 1.72)



| Location | 8,618,668 – 8,618,758 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 80.66 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -15.61 |
| Energy contribution | -17.55 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8618668 90 + 20766785 AACUGAGUGUCCUAUAUACGACAGA---------------UGACAGAUUUUCCACAAAUAUCAACAGCCUGGCCCUCAUUCGUGGGCCAUGGUUA----CUCAUAUGUU ...((((((((........)))...---------------.........................((((((((((........)))))).)))))----))))...... ( -23.40) >DroSec_CAF1 20958 108 + 1 AAGUGAGUGUCCUAUAUACGAAGAAUCUCGCUCACUGAG-UUGCGGAUUUUCCGCAAAUAUCAACAGCCUGGCUCUCAUUCGUGAGCCAUGGUUACUCUCUCAUAUAUU .((((((((...................))))))))(((-((((((.....))))))........((((((((((........)))))).)))).....)))....... ( -35.91) >DroSim_CAF1 18248 104 + 1 AACUGAGUGUCCUAUAUACGACGAUUCUCGCACACUGAG-UUGCGGAUUUUCCGCAAAUAUCAACAGCCUGGCUAUCAUUCGUGGGCCAUGGUUC----CUCAUAUAUU ...((((((((........))))............(((.-((((((.....))))))...)))..(((((((((..........))))).)))).----))))...... ( -25.70) >DroYak_CAF1 14465 100 + 1 AACUGAGUGUCCUAUAUACGAAG---CUCG--CACUCAGUUCACAGAUUUUCCGCAAAUAUCAACAAACUGGCUCUCAUUCGUCGGCCAUGGUUA----CUCAUAUAUU ((((((((((.((........))---...)--))))))))).....................(((....(((((..(....)..)))))..))).----.......... ( -18.50) >consensus AACUGAGUGUCCUAUAUACGAAGAA_CUCG__CACUGAG_UUACAGAUUUUCCGCAAAUAUCAACAGCCUGGCUCUCAUUCGUGGGCCAUGGUUA____CUCAUAUAUU ...((((.................................((((((.....))))))........((((((((((.(....).)))))).)))).....))))...... (-15.61 = -17.55 + 1.94)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:46 2006