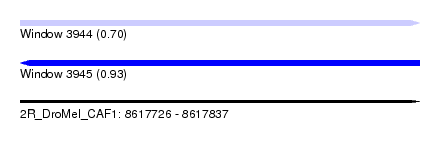
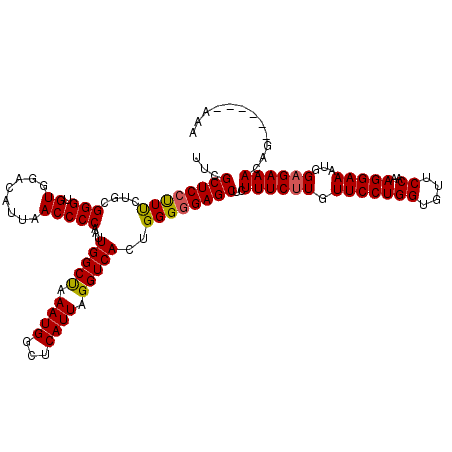
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,617,726 – 8,617,837 |
| Length | 111 |
| Max. P | 0.933717 |

| Location | 8,617,726 – 8,617,837 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -31.56 |
| Consensus MFE | -25.92 |
| Energy contribution | -25.84 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8617726 111 + 20766785 ---------CUGUUUCUCCAUUUCCUUUGGAACACCAGGAACAAGAAAUAGCUCCCCCAGUGACCUAAUGAGCCAUUUAGCCAAUUGGGGUUAAUGUCCACACCCGCAGAAGGGAGCGAA ---------.(((((((....(((((..((....)))))))..)))))))((((((...(((.((((((..((......))..)))))).....((....))..)))....))))))... ( -30.30) >DroSec_CAF1 20020 114 + 1 CUU------CUGUUUCUCCAUUUCCUUUGGAACACCAGGAACAAGAAACAGCUCCCCCAGUGACCUAAUGAGCCAUUUAGCCAAUUGGGGUUAAUGUCCACACCCGCAGAAAGGAGCGAU ...------.(((((((....(((((..((....)))))))..)))))))(((((((((((...((((........))))...)))))).....(((........)))....)))))... ( -30.50) >DroSim_CAF1 17313 114 + 1 CUU------CUGUUUCUCCAUUUCCUUUGGAACACCAGGAACAAGAAACAGCUCCCCCAGUGACCUAAUGAGCCAUUUAGCCAAUUGGGGUUAAUGUCCACACCCGCAGAAAGGAGCGAU ...------.(((((((....(((((..((....)))))))..)))))))(((((((((((...((((........))))...)))))).....(((........)))....)))))... ( -30.50) >DroEre_CAF1 17772 114 + 1 UUU------CUGUUUCUCCAUUUCCUUUGGAGCUCCAGGAACACGAAAGUGCUCUCCCACUGACCUAAUGAGCCAUUUGGCCAAUUGGGGUUAAUGUCCACACCCGCAGGAAGGAGCGAA (((------(((...(((((.......)))))...))))))(((....)))(.((((..(((.((((((..(((....)))..)))))).....((....))....)))...)))).).. ( -35.50) >DroYak_CAF1 13518 120 + 1 UUUCUGGUUCUGUUUCUCCAUUUCCUUUGGAACUCCAGGAACAAGAAAGAGCUCCCCCACUGACCUAAUGAGCCAUUUAGCCAAUUGGGGUUAAUGUCCACACCCGCAGAAAGGAGCGAA (((((.(((((.....((((.......))))......))))).)))))..(((((....(((......((..(...((((((......)))))).)..))......)))...)))))... ( -31.00) >consensus CUU______CUGUUUCUCCAUUUCCUUUGGAACACCAGGAACAAGAAACAGCUCCCCCAGUGACCUAAUGAGCCAUUUAGCCAAUUGGGGUUAAUGUCCACACCCGCAGAAAGGAGCGAA ..........(((((((....((((...))))....))))))).......(((((((((((...((((........))))...)))))).....(((........)))....)))))... (-25.92 = -25.84 + -0.08)

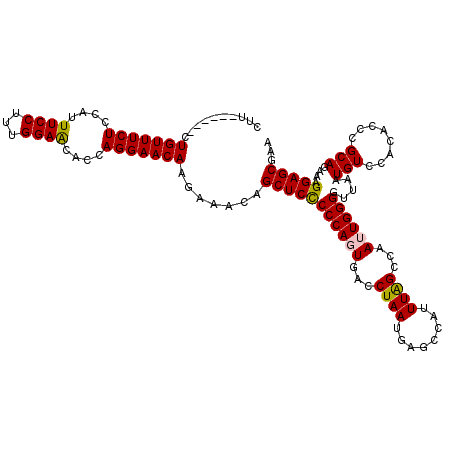
| Location | 8,617,726 – 8,617,837 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.92 |
| Mean single sequence MFE | -37.98 |
| Consensus MFE | -31.68 |
| Energy contribution | -31.60 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933717 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8617726 111 - 20766785 UUCGCUCCCUUCUGCGGGUGUGGACAUUAACCCCAAUUGGCUAAAUGGCUCAUUAGGUCACUGGGGGAGCUAUUUCUUGUUCCUGGUGUUCCAAAGGAAAUGGAGAAACAG--------- ...((((((((....(((.((........)))))...(((((.((((...)))).)))))..))))))))..((((((.(((((((....))..)))))...))))))...--------- ( -34.50) >DroSec_CAF1 20020 114 - 1 AUCGCUCCUUUCUGCGGGUGUGGACAUUAACCCCAAUUGGCUAAAUGGCUCAUUAGGUCACUGGGGGAGCUGUUUCUUGUUCCUGGUGUUCCAAAGGAAAUGGAGAAACAG------AAG .((((.(((......))).)))).......(((((..(((((.((((...)))).))))).)))))...(((((((((.(((((((....))..)))))...)))))))))------... ( -38.30) >DroSim_CAF1 17313 114 - 1 AUCGCUCCUUUCUGCGGGUGUGGACAUUAACCCCAAUUGGCUAAAUGGCUCAUUAGGUCACUGGGGGAGCUGUUUCUUGUUCCUGGUGUUCCAAAGGAAAUGGAGAAACAG------AAG .((((.(((......))).)))).......(((((..(((((.((((...)))).))))).)))))...(((((((((.(((((((....))..)))))...)))))))))------... ( -38.30) >DroEre_CAF1 17772 114 - 1 UUCGCUCCUUCCUGCGGGUGUGGACAUUAACCCCAAUUGGCCAAAUGGCUCAUUAGGUCAGUGGGAGAGCACUUUCGUGUUCCUGGAGCUCCAAAGGAAAUGGAGAAACAG------AAA ((((((((.(((..((((.((........)))))...(((((.((((...)))).))))))..)))((((((....))))))..)))))((((.......)))))))....------... ( -40.60) >DroYak_CAF1 13518 120 - 1 UUCGCUCCUUUCUGCGGGUGUGGACAUUAACCCCAAUUGGCUAAAUGGCUCAUUAGGUCAGUGGGGGAGCUCUUUCUUGUUCCUGGAGUUCCAAAGGAAAUGGAGAAACAGAACCAGAAA ...(((((..((..(....)..)).......((((.((((((.((((...)))).)))))))))))))))...((((.((((.((...(((((.......)))))...)))))).)))). ( -38.20) >consensus UUCGCUCCUUUCUGCGGGUGUGGACAUUAACCCCAAUUGGCUAAAUGGCUCAUUAGGUCACUGGGGGAGCUCUUUCUUGUUCCUGGUGUUCCAAAGGAAAUGGAGAAACAG______AAA ...((((((((....(((.((........)))))...(((((.((((...)))).)))))..))))))))..((((((.(((((((....))..)))))...))))))............ (-31.68 = -31.60 + -0.08)

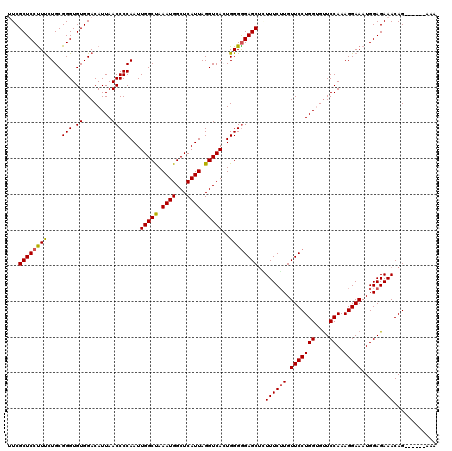
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:44 2006