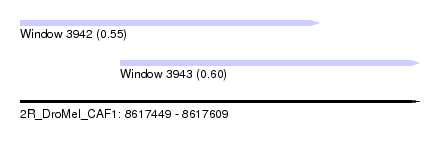
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,617,449 – 8,617,609 |
| Length | 160 |
| Max. P | 0.596806 |

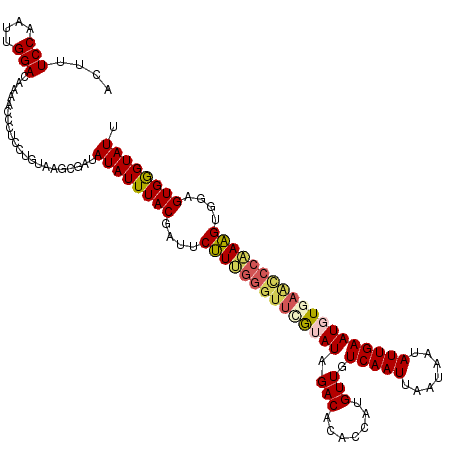
| Location | 8,617,449 – 8,617,569 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.31 |
| Mean single sequence MFE | -29.19 |
| Consensus MFE | -21.30 |
| Energy contribution | -22.10 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8617449 120 + 20766785 ACUUUCCAAAUGGACAAAACCCUCCUGUAAGCGAUAUAUUUACGAUUCUUUGGGUUUGUAUAGACACACCAUGUUGUCAAUUAAUAAUAUUGAAUGUGAACCCAAAGUGGAGUGAGUAUU ....(((....))).....................((((((((....(((((((((..(((.((((.....)))).(((((.......))))))))..)))))))))....)))))))). ( -29.00) >DroSec_CAF1 19743 120 + 1 ACUUUCCAAUUGGACAAAACCCUCCUGUAAGCGAUAUAUUUACGAUUCUUUGGGUUCGUAUAGACACACCAUGUUGUCAAUUAAUAAUAUUGAAUGUGAACCCAAAGUGGAGUGGGUAUU ....(((....))).....((((((((((((.......))))))...((((((((((((((.((((.....)))).(((((.......))))))))))))))))))).)))).))..... ( -31.50) >DroSim_CAF1 17036 120 + 1 ACUUUCCAAUUGGACAAAACCCUCCUGUAAGCGAAAUAUUUACGAUUCUUUGGGUUCGUAUAGACACACCAUGUUGUCAAUUAAUAAUAUUGAAUGUGAACCCAAAGUGGAGUGGGUAUU ....(((....))).....((((((((((((.......))))))...((((((((((((((.((((.....)))).(((((.......))))))))))))))))))).)))).))..... ( -31.50) >DroEre_CAF1 17504 111 + 1 ACUUUCCAAUUGGACAAAACC---CUGUAAGCGAUAUAUUUACGAUUCUUCGGGUUCGGAUGGACACACCGCGUUGUCAAUUAAUAAUAUUGAAUGU------GAAGUGGAGUGGGUAUU ....(((....)))....(((---(.....((..(((((((((((..(....)..)))(((.((((........)))).)))........)))))))------)..)).....))))... ( -23.70) >DroYak_CAF1 13244 116 + 1 ACUUUCCAAUUGGACAGAAGC---CUGUAAGCGAUAUAUUUACG-UUCCUAGGGUUCAGAUAGACACACCACGUUCUCAAUUAAUAAUAUUGAAUGUAAGUCCGAGGUGGAGUGGGUAUU .(.(((((.((((((.(((.(---(((..((((.((....))))-))..)))).))).............((((((...............))))))..))))))..))))).)...... ( -30.26) >consensus ACUUUCCAAUUGGACAAAACCCUCCUGUAAGCGAUAUAUUUACGAUUCUUUGGGUUCGUAUAGACACACCAUGUUGUCAAUUAAUAAUAUUGAAUGUGAACCCAAAGUGGAGUGGGUAUU ....(((....))).....................((((((((....((((((((((((((.(((.......))).(((((.......)))))))))))))))))))....)))))))). (-21.30 = -22.10 + 0.80)

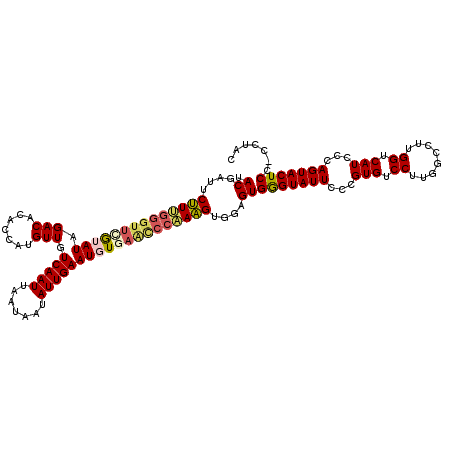
| Location | 8,617,489 – 8,617,609 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.24 |
| Mean single sequence MFE | -34.11 |
| Consensus MFE | -24.46 |
| Energy contribution | -25.26 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.596806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8617489 120 + 20766785 UACGAUUCUUUGGGUUUGUAUAGACACACCAUGUUGUCAAUUAAUAAUAUUGAAUGUGAACCCAAAGUGGAGUGAGUAUUGCCGUGUCCUUGGCCUUGGUCAUCCCAGUACUCCCCCCAC .......(((((((((..(((.((((.....)))).(((((.......))))))))..)))))))))(((.(.((((((((..(((.((........)).)))..)))))))).).))). ( -38.30) >DroSec_CAF1 19783 120 + 1 UACGAUUCUUUGGGUUCGUAUAGACACACCAUGUUGUCAAUUAAUAAUAUUGAAUGUGAACCCAAAGUGGAGUGGGUAUUCCCGUGUCCUUGGCCUUGGUCAUCCCAGUACUCUACCUAC ........(((((((((((((.((((.....)))).(((((.......))))))))))))))))))((((((((((....)))(((.((........)).)))......))))))).... ( -33.80) >DroSim_CAF1 17076 120 + 1 UACGAUUCUUUGGGUUCGUAUAGACACACCAUGUUGUCAAUUAAUAAUAUUGAAUGUGAACCCAAAGUGGAGUGGGUAUUCCCGUGUCCUUGGCCUUGGUCAUCCCAGUACUCCACCUAC ........(((((((((((((.((((.....)))).(((((.......))))))))))))))))))((((((((((....)))(((.((........)).)))......))))))).... ( -36.10) >DroEre_CAF1 17541 112 + 1 UACGAUUCUUCGGGUUCGGAUGGACACACCGCGUUGUCAAUUAAUAAUAUUGAAUGU------GAAGUGGAGUGGGUAUUCCUGUGUCCUUGGCCUUGGUCAUC-CAGUACUCC-CUUAC (((((..(....)..))(((((((((((.(((((..(((((.......)))))))))------)....(((((....))))))))))))..(((....))))))-).)))....-..... ( -32.20) >DroYak_CAF1 13281 117 + 1 UACG-UUCCUAGGGUUCAGAUAGACACACCACGUUCUCAAUUAAUAAUAUUGAAUGUAAGUCCGAGGUGGAGUGGGUAUUCCCGUGUCCUUGGCCUUGGUCAUCCCAGUACUCC-C-UAC ....-....(((((................((((((...............))))))..(.(((((((.(((((((....))))....))).))))))).)...........))-)-)). ( -30.16) >consensus UACGAUUCUUUGGGUUCGUAUAGACACACCAUGUUGUCAAUUAAUAAUAUUGAAUGUGAACCCAAAGUGGAGUGGGUAUUCCCGUGUCCUUGGCCUUGGUCAUCCCAGUACUCC_CCUAC .((....((((((((((((((.(((.......))).(((((.......)))))))))))))))))))....))(((((((...(((.((........)).)))...)))))))....... (-24.46 = -25.26 + 0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:42 2006