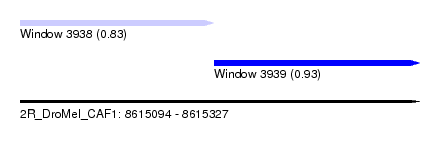
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,615,094 – 8,615,327 |
| Length | 233 |
| Max. P | 0.930058 |

| Location | 8,615,094 – 8,615,207 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.80 |
| Mean single sequence MFE | -33.26 |
| Consensus MFE | -23.09 |
| Energy contribution | -23.73 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828550 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8615094 113 + 20766785 ---GGCAGGCCAAG-GGGUGAGAUGCCAAAAAGCGACUGUGAAAUAUUCGCAGCUGACAGUUUUGAAAAAUGAAAGCUGAGUGGGCGGUGCACUGGGU---UGGUGGGCGGAGGGCGGAG ---.((..(((((.-.((((...((((......((.((((((.....)))))).)).(((((((........)))))))....))))...))))...)---))))..))........... ( -33.50) >DroSec_CAF1 17434 106 + 1 ---GGCAGGCCAAG-GGGUGAGAUGCCAAAAAGCGACUGUGAAAUAUUCGCAGCUGACAGUUUUGAAAAAUGAAAGCUGUGUGGGCGGUGCACUGCGU---UG-------GAGGGCGGAG ---.((...(((((-.((((...((((......((.((((((.....)))))).))((((((((........))))))))...))))...)))).).)---))-------)...)).... ( -35.80) >DroSim_CAF1 14033 106 + 1 ---GGCAGGCCAAG-GGGUGAGAUGCCAAAAAGCGACUGUGAAAUAUUCGCAGCUGACAGUUUUGAAAAAUGAAAGCUGUGUGGGCGGUGCACUGCGU---UG-------GAGGGCGGAG ---.((...(((((-.((((...((((......((.((((((.....)))))).))((((((((........))))))))...))))...)))).).)---))-------)...)).... ( -35.80) >DroEre_CAF1 15263 100 + 1 ---GGCGGGCCAGG-GGCUGAGAUGCCAAAAAGCGACUGUGAAAUAUUCGCAGCUGACAGUUUUGAAAAAUGAAAGCUGAGAGGGCGGUG----------------GGCGGUGGGUGGUG ---.((.(.(((..-(.(((...((((......((.((((((.....)))))).)).(((((((........)))))))....)))).))----------------).)..))).).)). ( -29.20) >DroYak_CAF1 10831 120 + 1 AAUGGAAGGCCAGGGGGGUGAGAUGCCAAAAAGCGACUGUGAAAUAUUCGCAGCUGACAGUUUUGAAAAAUGAAAGCUGAGAGGGCGGUGCACUUGGAGCACGGAGGACGGAGGGCGGUA ........(((....(((((...((((......((.((((((.....)))))).)).(((((((........)))))))....))))...)))))...((.((.....))....))))). ( -32.00) >consensus ___GGCAGGCCAAG_GGGUGAGAUGCCAAAAAGCGACUGUGAAAUAUUCGCAGCUGACAGUUUUGAAAAAUGAAAGCUGAGUGGGCGGUGCACUGCGU___UG___G_CGGAGGGCGGAG ........(((.....((((...((((......((.((((((.....)))))).)).(((((((........)))))))....))))...))))...................))).... (-23.09 = -23.73 + 0.64)


| Location | 8,615,207 – 8,615,327 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.83 |
| Mean single sequence MFE | -41.52 |
| Consensus MFE | -37.70 |
| Energy contribution | -38.34 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930058 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8615207 120 + 20766785 GGCGUGGCACAAGCCAUAAAGAUCCAGUGGCAGCUUCAGCGGAAGCGGAAGCGUUCGGCUCGUGCGAAUAUUGCAAAUGUGGCGGAAAAUAUGGCGUCAGUCAAAAGUCAGCGCAGCGCG (((.((((....((((((....(((.((.((((((((.((....)).)))))(((((.(....))))))........))).)))))...))))))....))))...))).(((...))). ( -43.20) >DroSec_CAF1 17540 120 + 1 GGCGUGGCACAAGCCAUAAAGAUCCAGUGGCAGUUUCAGCGGAAGCGGAAGCGUUCGGCUCGUGCGAAUAUUGCAAAUAUGGCGGAAAAUAUGGCGUCAGUCAAAAGUCAGCGCAGCGCG .((((.((.(..((((((..((.((...(((.(((((.((....)).)))))))).)).)).(((((...)))))..)))))).((.....((((....))))....)).).)).)))). ( -38.90) >DroSim_CAF1 14139 120 + 1 GGCGUGGCACAAGCCAUAAAGAUCCAGUGGCAGUUUCAGCGGAAGCGGAAGCGUUCGGCUCGUGCGAAUAUUGCAAAUGUGGCGGAAAAUAUGGCGUCAGUCAAAAGUCAGCGCAGCGCG (((.((((....((((((....(((.((.((((((((.((....)).)))))(((((.(....))))))........))).)))))...))))))....))))...))).(((...))). ( -40.80) >DroEre_CAF1 15363 114 + 1 GGCGUGGCGCAAGCCAUAAAGAUCCAGUGGCAGUUUCAGCGG------AAGCGUUCGGCUCGUGCGAAUAUUGCAAAUGUGGCGGAAAAUAUGGCGUCAGUCAAAAGUCAGCGCAGCGCG .((((.((((..((((((....(((.((.(((.((.((((..------..))(((((.(....))))))..)).)).))).)))))...))))))(.(........).).)))).)))). ( -39.40) >DroYak_CAF1 10951 120 + 1 GGCGUGGCGCAAGCCAUAAAGAUCCAGUGGCAGUUUCAGCGGAAGCGGAAGCGUUCGGCUCGUGCGAAUAUUGCAAAUGUGGCGGAAAAUAUGGCGUCAGUCAAAAGUCAGCGCAGCGCG .((((.((((..((((((....(((.((.((((((((.((....)).)))))(((((.(....))))))........))).)))))...))))))(.(........).).)))).)))). ( -45.30) >consensus GGCGUGGCACAAGCCAUAAAGAUCCAGUGGCAGUUUCAGCGGAAGCGGAAGCGUUCGGCUCGUGCGAAUAUUGCAAAUGUGGCGGAAAAUAUGGCGUCAGUCAAAAGUCAGCGCAGCGCG (((.((((....((((((....(((.((.((((((((.((....)).)))))(((((.(....))))))........))).)))))...))))))....))))...))).(((...))). (-37.70 = -38.34 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:38 2006