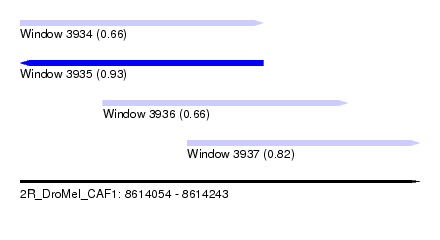
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,614,054 – 8,614,243 |
| Length | 189 |
| Max. P | 0.926992 |

| Location | 8,614,054 – 8,614,169 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.14 |
| Mean single sequence MFE | -34.05 |
| Consensus MFE | -22.56 |
| Energy contribution | -24.38 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8614054 115 + 20766785 UGGCCGACAUCUCUUUAUCGUGUGAUUCAUUCUGGUU-UGCCUUUGGCAUUGGCUGUGACUCUGGCUCUGGGUCUGGAUUUUCCAGCCCUUUCCCCAACCGAGAUCC----CUGGCCCAC .(((((..(((((..........((.((((.(..(..-((((...)))))..)..)))).))(((....(((.((((.....))))))).....)))...)))))..----.)))))... ( -35.70) >DroSec_CAF1 16382 115 + 1 UGGCCGACAUCUCUUUAUCGUGUGAUUCAUUCUGGUU-UGCCUCUGGCAUUGGCUGUGACUCUGGCUCUGGGUCUGGAUUUUCCAGGCCUUACCCAAACCAAGAUCC----CUGGCCCAC .((((((.(((.(........).)))))((..(((((-((((.........))).........((....((((((((.....))))))))...))))))))..))..----..))))... ( -35.30) >DroSim_CAF1 12616 109 + 1 UGGCCGACAUCUCUUUAUCGUGUGAUUCAUUCUGGUU-UGCCUCUGGCAUUGGCUGUGACUC------UGGGUCUGGAUUUUCCAGGCCUUACCCAAACCAAGAUCC----CUGGCCCAC .((((((.(((.(........).)))))((..(((((-((..((((((....)))).))...------.((((((((.....))))))))....)))))))..))..----..))))... ( -33.40) >DroEre_CAF1 14214 97 + 1 UGGCCGACAUCUCUUUAUCGUGUGAUUCAUUCGGGUUUUGCCUGUGGCAUUGGCUCCGUCCCCGGCUUUG-----------------------CCAUACCGAGAUCCAGAUCUGGCCCAC .((((((.(((.(........).)))))....((((((((..(((((((..((((........)))).))-----------------------))))).))))))))......))))... ( -31.80) >consensus UGGCCGACAUCUCUUUAUCGUGUGAUUCAUUCUGGUU_UGCCUCUGGCAUUGGCUGUGACUCUGGCUCUGGGUCUGGAUUUUCCAGGCCUUACCCAAACCAAGAUCC____CUGGCCCAC .((((((.(((.(........).)))))(((.(((((..(((.........))).........((....((((((((.....))))))))...)).))))).)))........))))... (-22.56 = -24.38 + 1.81)

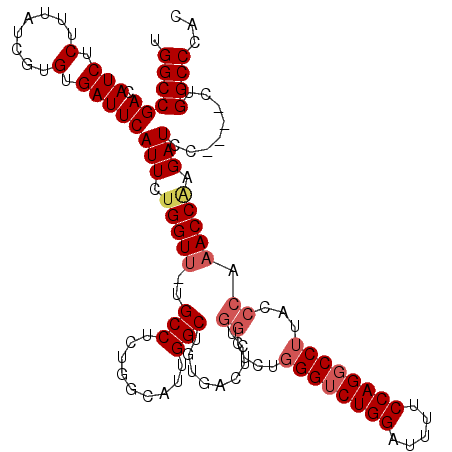
| Location | 8,614,054 – 8,614,169 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.14 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -23.10 |
| Energy contribution | -23.97 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8614054 115 - 20766785 GUGGGCCAG----GGAUCUCGGUUGGGGAAAGGGCUGGAAAAUCCAGACCCAGAGCCAGAGUCACAGCCAAUGCCAAAGGCA-AACCAGAAUGAAUCACACGAUAAAGAGAUGUCGGCCA ...((((.(----(.((((((((((.((...(((((((.....)))).)))....)).......)))))..((((...))))-........................))))).)))))). ( -40.50) >DroSec_CAF1 16382 115 - 1 GUGGGCCAG----GGAUCUUGGUUUGGGUAAGGCCUGGAAAAUCCAGACCCAGAGCCAGAGUCACAGCCAAUGCCAGAGGCA-AACCAGAAUGAAUCACACGAUAAAGAGAUGUCGGCCA ...((((.(----((((..((((((((((.....((((.....))))))))))).)))..)))...(((.........))).-..))..............((((......)))))))). ( -37.60) >DroSim_CAF1 12616 109 - 1 GUGGGCCAG----GGAUCUUGGUUUGGGUAAGGCCUGGAAAAUCCAGACCCA------GAGUCACAGCCAAUGCCAGAGGCA-AACCAGAAUGAAUCACACGAUAAAGAGAUGUCGGCCA ...((((.(----(.((((((((((((((.....((((.....)))))))).------........(((.........))))-)))).......(((....)))...))))).)))))). ( -33.50) >DroEre_CAF1 14214 97 - 1 GUGGGCCAGAUCUGGAUCUCGGUAUGG-----------------------CAAAGCCGGGGACGGAGCCAAUGCCACAGGCAAAACCCGAAUGAAUCACACGAUAAAGAGAUGUCGGCCA ...((((.(((....((((((((.(((-----------------------(....(((....))).)))).((((...))))..))).......(((....)))...)))))))))))). ( -33.60) >consensus GUGGGCCAG____GGAUCUCGGUUUGGGUAAGGCCUGGAAAAUCCAGACCCAGAGCCAGAGUCACAGCCAAUGCCAGAGGCA_AACCAGAAUGAAUCACACGAUAAAGAGAUGUCGGCCA ...((((.......(((.(((.(((((....((.((((.....)))).))................(((.........)))....))))).))))))....((((......)))))))). (-23.10 = -23.97 + 0.87)

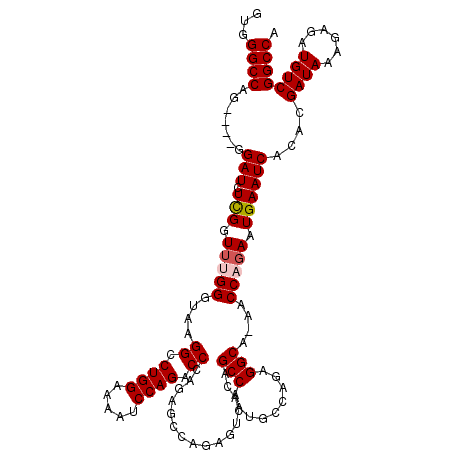
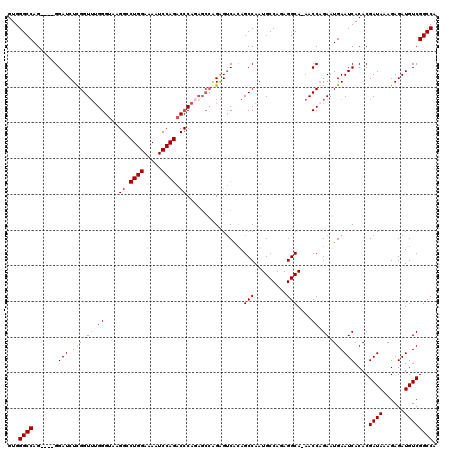
| Location | 8,614,093 – 8,614,209 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -19.34 |
| Energy contribution | -21.03 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8614093 116 + 20766785 CCUUUGGCAUUGGCUGUGACUCUGGCUCUGGGUCUGGAUUUUCCAGCCCUUUCCCCAACCGAGAUCC----CUGGCCCACUUUAUCUCAAGUUUAUCUCAAUUUGAGGAAAGAAACCUCG .....((....(((((.((((((((....(((.((((.....))))))).....)))...))).)).----).))))..((((.(((((((((......)))))))))))))...))... ( -32.10) >DroSec_CAF1 16421 114 + 1 CCUCUGGCAUUGGCUGUGACUCUGGCUCUGGGUCUGGAUUUUCCAGGCCUUACCCAAACCAAGAUCC----CUGGCCCACUUUAUCUCAAGUUUAUCUCAAUUUGAGGAAAGAAACCU-- .....(((...((.(.(.....(((....((((((((.....))))))))...))).....).).))----...)))..((((.(((((((((......)))))))))))))......-- ( -32.10) >DroSim_CAF1 12655 110 + 1 CCUCUGGCAUUGGCUGUGACUC------UGGGUCUGGAUUUUCCAGGCCUUACCCAAACCAAGAUCC----CUGGCCCACUUUAUCUCAAGUUUAUCUCAAUUUGAGGAAAGAAACCUCG .....(((.((((...((....------.((((((((.....))))))))....))..)))).....----...)))..((((.(((((((((......)))))))))))))........ ( -30.20) >DroEre_CAF1 14254 97 + 1 CCUGUGGCAUUGGCUCCGUCCCCGGCUUUG-----------------------CCAUACCGAGAUCCAGAUCUGGCCCACUUUAACUCAAGUUUAUCUCAAUUUGAGGAAAGAAACCUCG ..(((((((..((((........)))).))-----------------------))))).((((..(((....)))....((((..((((((((......)))))))).))))....)))) ( -26.40) >consensus CCUCUGGCAUUGGCUGUGACUCUGGCUCUGGGUCUGGAUUUUCCAGGCCUUACCCAAACCAAGAUCC____CUGGCCCACUUUAUCUCAAGUUUAUCUCAAUUUGAGGAAAGAAACCUCG .....((....((((........((....((((((((.....))))))))...))......((........))))))..((((.(((((((((......)))))))))))))...))... (-19.34 = -21.03 + 1.69)


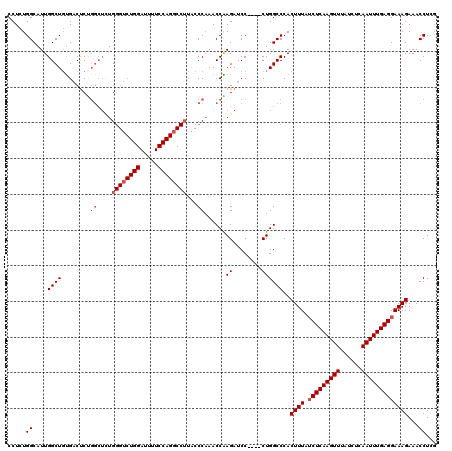
| Location | 8,614,133 – 8,614,243 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.25 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -22.66 |
| Energy contribution | -22.86 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8614133 110 + 20766785 UUCCAGCCCUUUCCCCAACCGAGAUCC----CUGGCCCACUUUAUCUCAAGUUUAUCUCAAUUUGAGGAAAGAAACCUCGGUCUCCG---GAGCAUUUGGCAUGCAGU---GCUCGGGCU ....(((((.........(.((((((.----..((....((((.(((((((((......)))))))))))))...))..)))))).)---(((((((........)))---))))))))) ( -35.20) >DroSec_CAF1 16461 104 + 1 UUCCAGGCCUUACCCAAACCAAGAUCC----CUGGCCCACUUUAUCUCAAGUUUAUCUCAAUUUGAGGAAAGAAACCU------CCG---GAGCAUUUGGCAUGCAGU---GCUCGGGCU ...((((.(((.........)))...)----)))((((.((((.(((((((((......)))))))))))))......------...---(((((((........)))---)))))))). ( -29.70) >DroSim_CAF1 12689 110 + 1 UUCCAGGCCUUACCCAAACCAAGAUCC----CUGGCCCACUUUAUCUCAAGUUUAUCUCAAUUUGAGGAAAGAAACCUCGGUCUCCG---GAGCAUUUGGCAUGCAGU---GCUCGGGCU ...((((.(((.........)))...)----)))((((.((((.(((((((((......)))))))))))))......(((...)))---(((((((........)))---)))))))). ( -30.70) >DroEre_CAF1 14284 104 + 1 -------------CCAUACCGAGAUCCAGAUCUGGCCCACUUUAACUCAAGUUUAUCUCAAUUUGAGGAAAGAAACCUCGGUCUCCG---GAGCAUUGGGCAUGCACUCGGGCUCGGGCU -------------.....(((((.(((((.....(((((((((..((((((((......)))))))).))))...(..(((...)))---..)...))))).....)).))))))))... ( -32.70) >DroYak_CAF1 9841 94 + 1 -------------------AGAGAUCC----CUGGCCCACUUUAUCUCAAGUUUAUCUCAAUUUGAGGAAAGAAACCUCGGUCUCCGCAGGAGCAUUUGGCAUGCACU---GCUCGGGCU -------------------........----..(((((.((((.(((((((((......)))))))))))))..............((((..((((.....)))).))---))..))))) ( -28.20) >consensus UUCCAG_CCUU_CCCAAACCGAGAUCC____CUGGCCCACUUUAUCUCAAGUUUAUCUCAAUUUGAGGAAAGAAACCUCGGUCUCCG___GAGCAUUUGGCAUGCAGU___GCUCGGGCU .................................(((((.((((.(((((((((......)))))))))))))..................((((.....(....)......))))))))) (-22.66 = -22.86 + 0.20)

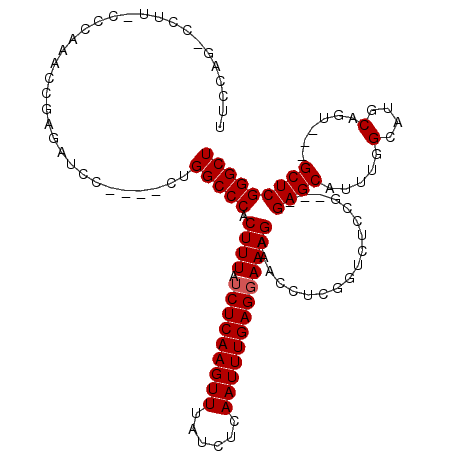
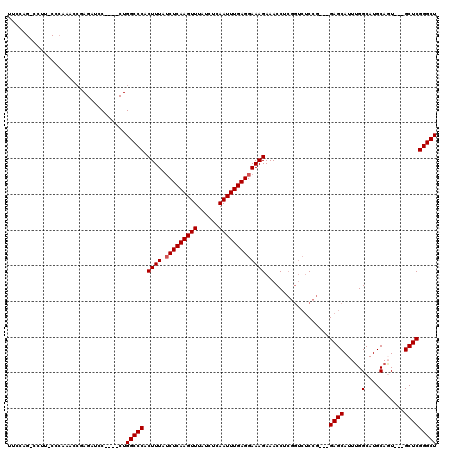
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:36 2006