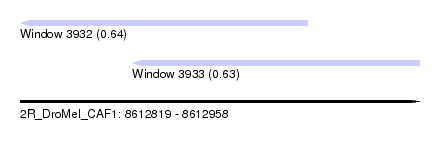
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,612,819 – 8,612,958 |
| Length | 139 |
| Max. P | 0.636181 |

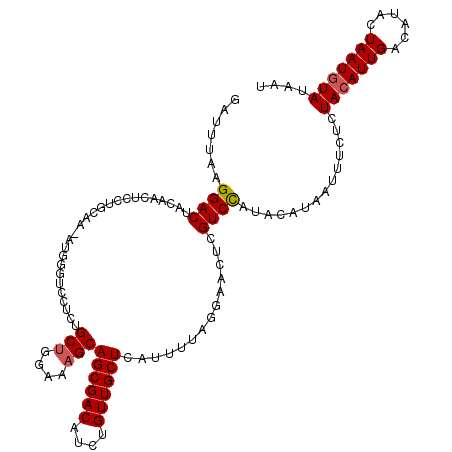
| Location | 8,612,819 – 8,612,919 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.23 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -16.82 |
| Energy contribution | -17.27 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8612819 100 - 20766785 AAUUUAAGGAC-------------------GUCCUCUGCUGGAAAGCAGCGACAUCUGUUGCUCAUUUUUGGAACUUGUCUAUACAUAAUUUCUCUACAUUGACUUACUAAUCUAUAAG .......((((-------------------((((..((((....))))(((((....)))))........)))...)))))...................................... ( -17.90) >DroSec_CAF1 15137 119 - 1 GAUUCAAGGACUACAACUCCUGCAAGAUGGGUCGUCAGCUGGAAUGCAGCGACAUCUGUUGCUCAUUUUAGGAACUCGUCCAUACAUAAUUUCUCUACAUUGACAUACUAAUGUAGAAU .......((((.((.((.((........)))).))...((((((((.((((((....))))))))))))))......))))............(((((((((......))))))))).. ( -33.30) >DroSim_CAF1 11372 119 - 1 GAUUUAAGGACUACAACUCCUGCAAAAUGGGUUCACUGCUGGAAAGCAGCGACAUCUGUUGCUCAUUUUAGGAACUCGUCCAUACAUAAUUUCUCUACAUUGACAUACUAAUGUAUAAU .......((((......(((((..((((((((((.(((((....))))).))........)))))))))))))....))))..............(((((((......))))))).... ( -29.70) >consensus GAUUUAAGGACUACAACUCCUGCAA_AUGGGUCCUCUGCUGGAAAGCAGCGACAUCUGUUGCUCAUUUUAGGAACUCGUCCAUACAUAAUUUCUCUACAUUGACAUACUAAUGUAUAAU .......((((..........................(((....)))((((((....))))))..............))))..............(((((((......))))))).... (-16.82 = -17.27 + 0.45)

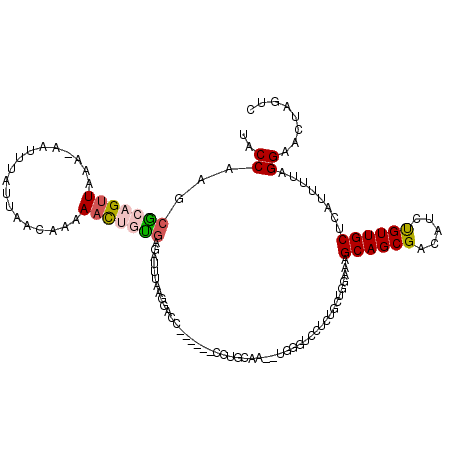
| Location | 8,612,858 – 8,612,958 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.73 |
| Mean single sequence MFE | -25.65 |
| Consensus MFE | -10.01 |
| Energy contribution | -10.41 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.39 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8612858 100 - 20766785 UACCAAACGCAGUUAAA-AAUUUAUUAACAAAAACUGUGAAAUUUAAGGAC-------------------GUCCUCUGCUGGAAAGCAGCGACAUCUGUUGCUCAUUUUUGGAACUUGUC ..((((((((((((...-..............)))))))........(((.-------------------(((..(((((....))))).))).)))..........)))))........ ( -23.93) >DroSec_CAF1 15176 119 - 1 UACCAAGCACAGUUAAA-AAUUUAUUAACAAAACCUUUGAGAUUCAAGGACUACAACUCCUGCAAGAUGGGUCGUCAGCUGGAAUGCAGCGACAUCUGUUGCUCAUUUUAGGAACUCGUC ......(((..(((((.-......))))).....(((((.....)))))...........)))..(((((((..((..((((((((.((((((....))))))))))))))))))))))) ( -29.30) >DroSim_CAF1 11411 119 - 1 UGCCAAGCGCAGUUAAA-AAUUUAUUAACAAAAACUGUGAGAUUUAAGGACUACAACUCCUGCAAAAUGGGUUCACUGCUGGAAAGCAGCGACAUCUGUUGCUCAUUUUAGGAACUCGUC (((....(((((((...-..............))))))).......((((.......)))))))..((((((((.(((((....)))((((((....))))))......)))))))))). ( -30.73) >DroEre_CAF1 13044 89 - 1 UUCCAAGCGCAGUUCAA-AAAUAAUUAACAGAAAUCGCGAGGUUUAAGGACC----------------------------AUG--GCAGCGCCAUCUGUUGCACAUUAUGGGAACUAGUA ......((..(((((..-..((((((((((((........((((....))))----------------------------.((--((...)))))))))))...)))))..))))).)). ( -21.80) >DroYak_CAF1 8545 114 - 1 UACCAAACACUGUUAAAAAAAUUAUUAACCGAAAUCAUCAGAUUAAAGGUCC------CCUGCAAAUUGGGAUUUACACUAUGAAGCAGCGACAUCCGUUGCAUAUCUUUGGAACUAGUA ..(((((((..((((((..........(((..((((....))))...))).(------((........))).)))).))..))..((((((.....)))))).....)))))........ ( -22.50) >consensus UACCAAGCGCAGUUAAA_AAUUUAUUAACAAAAACUGUGAGAUUUAAGGACC______CCUGCAA__UGGGUCCUCUGCUGGAAAGCAGCGACAUCUGUUGCUCAUUUUAGGAACUAGUC ..((...(((((((..................)))))))..............................................((((((.....))))))........))........ (-10.01 = -10.41 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:32 2006