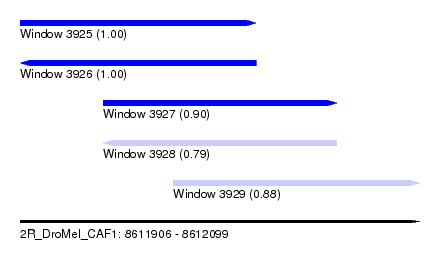
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,611,906 – 8,612,099 |
| Length | 193 |
| Max. P | 0.999876 |

| Location | 8,611,906 – 8,612,020 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -43.38 |
| Consensus MFE | -35.26 |
| Energy contribution | -35.18 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.64 |
| Structure conservation index | 0.81 |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998855 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8611906 114 + 20766785 ACUACUUGGCAUUCCAUUGGCAUGCGCCACAUUUCAGAAGGUCCGGCAGGUGUU------UUCCUGCUGAUUUAAAUUAUUUGCAUGCCUUACUGGGGUGUCGAGCCCCCGGCAUAAAUU .....(((((((((((..((((((((((...........))).(((((((....------..))))))).............)))))))....)))))))))))(((...)))....... ( -42.90) >DroSec_CAF1 12816 114 + 1 ACUACUUGGCAUUCCAUUGGCAUGCGGCACAUUUCAGAAGGUCCGGCAGGUGUU------UUCCUGCUGAUUUAAAUUAUUUGCAUGCCUUACUGGGGGGCCAUGCCCCCGGCAUAAAUU .......(((.(((((..((((((((((............)))(((((((....------..))))))).............)))))))....))))).)))(((((...)))))..... ( -40.60) >DroSim_CAF1 9674 114 + 1 ACUACUUGGCAUUCCAUUGGCAUGCGCCACAUUUCAGAAGGUCCGGCAGGUGUU------UUCCUGCUGAUUUAAAUUAUUUGCAUGCCUUACUGGGGGGCCAUGCCCCCGGCAUAAAUU .......(((.(((((..((((((((((...........))).(((((((....------..))))))).............)))))))....))))).)))(((((...)))))..... ( -41.40) >DroEre_CAF1 12146 119 + 1 GCUACUUGGCAUUCCAUUGGCAUGCGCCACAUUGCAGAAAGUCCGGCAGGUGUUUUCGUUUUCCUGCUGAUUUAAAUUAUUUGCAUGCCUUGCUGGGG-GCCAAGCCCCCGGCAUAAAUU (((....)))........(((((((((......))...(((((.((((((............))))))))))).........))))))).((((((((-((...))))))))))...... ( -48.00) >DroYak_CAF1 7625 120 + 1 ACUACUUGGCAUUCCAUUGGCAUGCGCCACAUUUCAAAAGGUCCGGCAGGUGUUUUCGUUUUCCUGCUGAUUUAAAUUAUUUGCAUGCCUUGCUGGGGGGCAAAGCCCCCGGCAUAAAUU ......(((....)))..((((((((((...........))).(((((((............))))))).............))))))).(((((((((......)))))))))...... ( -44.00) >consensus ACUACUUGGCAUUCCAUUGGCAUGCGCCACAUUUCAGAAGGUCCGGCAGGUGUU______UUCCUGCUGAUUUAAAUUAUUUGCAUGCCUUACUGGGGGGCCAAGCCCCCGGCAUAAAUU ......((((.(((((..((((((((....((((....(((((.((((((............)))))))))))))))....))))))))....))))).)))).(((...)))....... (-35.26 = -35.18 + -0.08)

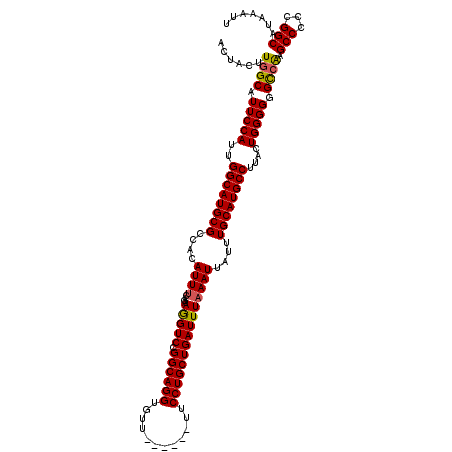
| Location | 8,611,906 – 8,612,020 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.64 |
| Mean single sequence MFE | -38.68 |
| Consensus MFE | -34.84 |
| Energy contribution | -34.44 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.34 |
| SVM RNA-class probability | 0.999876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8611906 114 - 20766785 AAUUUAUGCCGGGGGCUCGACACCCCAGUAAGGCAUGCAAAUAAUUUAAAUCAGCAGGAA------AACACCUGCCGGACCUUCUGAAAUGUGGCGCAUGCCAAUGGAAUGCCAAGUAGU ......(((.((((........)))).))).(((((((...............(((((..------....)))))(((.....))).........)))))))..(((....)))...... ( -35.80) >DroSec_CAF1 12816 114 - 1 AAUUUAUGCCGGGGGCAUGGCCCCCCAGUAAGGCAUGCAAAUAAUUUAAAUCAGCAGGAA------AACACCUGCCGGACCUUCUGAAAUGUGCCGCAUGCCAAUGGAAUGCCAAGUAGU ...((((...((((((...))))))..))))(((((((...............(((((..------....)))))(((.....))).........)))))))..(((....)))...... ( -38.50) >DroSim_CAF1 9674 114 - 1 AAUUUAUGCCGGGGGCAUGGCCCCCCAGUAAGGCAUGCAAAUAAUUUAAAUCAGCAGGAA------AACACCUGCCGGACCUUCUGAAAUGUGGCGCAUGCCAAUGGAAUGCCAAGUAGU ...((((...((((((...))))))..))))(((((((...............(((((..------....)))))(((.....))).........)))))))..(((....)))...... ( -38.50) >DroEre_CAF1 12146 119 - 1 AAUUUAUGCCGGGGGCUUGGC-CCCCAGCAAGGCAUGCAAAUAAUUUAAAUCAGCAGGAAAACGAAAACACCUGCCGGACUUUCUGCAAUGUGGCGCAUGCCAAUGGAAUGCCAAGUAGC ......(((.((((((...))-)))).))).(((((((............((.(((((............))))).)).......((......)))))))))..(((....)))...... ( -41.10) >DroYak_CAF1 7625 120 - 1 AAUUUAUGCCGGGGGCUUUGCCCCCCAGCAAGGCAUGCAAAUAAUUUAAAUCAGCAGGAAAACGAAAACACCUGCCGGACCUUUUGAAAUGUGGCGCAUGCCAAUGGAAUGCCAAGUAGU ......(((.((((((...))))))..))).(((((((..(((.((((((((.(((((............)))))..))...)))))).)))...)))))))..(((....)))...... ( -39.50) >consensus AAUUUAUGCCGGGGGCUUGGCCCCCCAGUAAGGCAUGCAAAUAAUUUAAAUCAGCAGGAA______AACACCUGCCGGACCUUCUGAAAUGUGGCGCAUGCCAAUGGAAUGCCAAGUAGU ......(((.((((........)))).))).(((((((...............(((((............)))))(((.....))).........)))))))..(((....)))...... (-34.84 = -34.44 + -0.40)

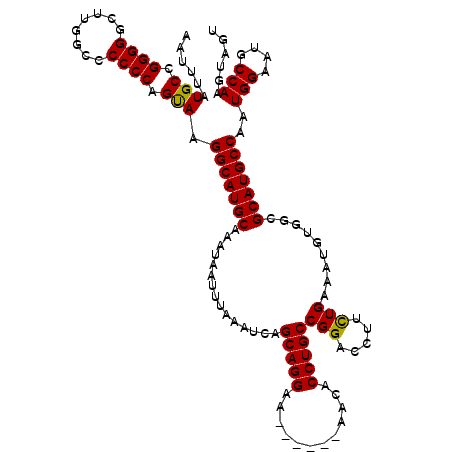
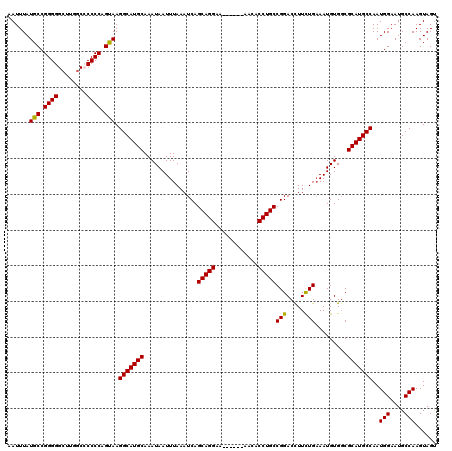
| Location | 8,611,946 – 8,612,059 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.43 |
| Mean single sequence MFE | -37.84 |
| Consensus MFE | -30.00 |
| Energy contribution | -30.28 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8611946 113 + 20766785 GUCCGGCAGGUGUU------UUCCUGCUGAUUUAAAUUAUUUGCAUGCCUUACUGGGGUGUCGAGCCCCCGGCAUAAAUUCC-CCCGCCGGCCAGAAAUUUUAACCAAAGCCAAUUGGCC ...(((((((....------..)))))))..(((((...((((...(((.....(((((.....))))).(((.........-...))))))))))...))))).....(((....))). ( -33.60) >DroSec_CAF1 12856 113 + 1 GUCCGGCAGGUGUU------UUCCUGCUGAUUUAAAUUAUUUGCAUGCCUUACUGGGGGGCCAUGCCCCCGGCAUAAAUUCC-CCCGCCGGCCAGAAAUUUUAACCAAAGCCAAUUGGCC ...(((((((....------..)))))))..(((((...((((...(((......((((((...))))))(((.........-...))))))))))...))))).....(((....))). ( -37.80) >DroSim_CAF1 9714 113 + 1 GUCCGGCAGGUGUU------UUCCUGCUGAUUUAAAUUAUUUGCAUGCCUUACUGGGGGGCCAUGCCCCCGGCAUAAAUUCC-CCCGCCGGCCAGAAAUUUUAACCAAAGCCAAUUGGCC ...(((((((....------..)))))))..(((((...((((...(((......((((((...))))))(((.........-...))))))))))...))))).....(((....))). ( -37.80) >DroEre_CAF1 12186 119 + 1 GUCCGGCAGGUGUUUUCGUUUUCCUGCUGAUUUAAAUUAUUUGCAUGCCUUGCUGGGG-GCCAAGCCCCCGGCAUAAAUUCCCCCCGGCGGCCGCACAUUUUAACCAAAGCCAAUUGGCC ...(((((((............)))))))............(((..(((.((((((((-((...))))))))))......((....)).))).))).............(((....))). ( -40.90) >DroYak_CAF1 7665 119 + 1 GUCCGGCAGGUGUUUUCGUUUUCCUGCUGAUUUAAAUUAUUUGCAUGCCUUGCUGGGGGGCAAAGCCCCCGGCAUAAAUUCC-CCCGGCGGCCAGAAAUUUUAACCAAAGCCAAUUGGCC ...(((((((............)))))))..(((((...((((...(((..(((((((((....(((...)))......)))-)))))))))))))...))))).....(((....))). ( -39.10) >consensus GUCCGGCAGGUGUU______UUCCUGCUGAUUUAAAUUAUUUGCAUGCCUUACUGGGGGGCCAAGCCCCCGGCAUAAAUUCC_CCCGCCGGCCAGAAAUUUUAACCAAAGCCAAUUGGCC ...(((((((............)))))))...............(((((......((((((...)))))))))))..............((((((...((((....))))....)))))) (-30.00 = -30.28 + 0.28)

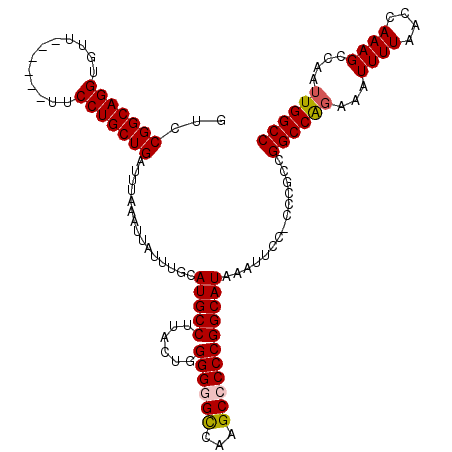
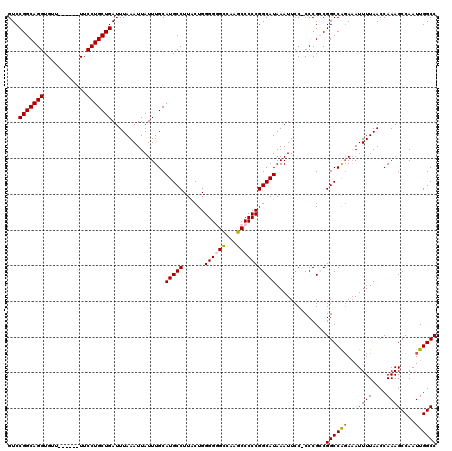
| Location | 8,611,946 – 8,612,059 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.43 |
| Mean single sequence MFE | -38.86 |
| Consensus MFE | -31.80 |
| Energy contribution | -32.20 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8611946 113 - 20766785 GGCCAAUUGGCUUUGGUUAAAAUUUCUGGCCGGCGGG-GGAAUUUAUGCCGGGGGCUCGACACCCCAGUAAGGCAUGCAAAUAAUUUAAAUCAGCAGGAA------AACACCUGCCGGAC ((((((......))))))...........((((((((-.......(((((.((((.......)))).....)))))((...............)).....------....)))))))).. ( -36.16) >DroSec_CAF1 12856 113 - 1 GGCCAAUUGGCUUUGGUUAAAAUUUCUGGCCGGCGGG-GGAAUUUAUGCCGGGGGCAUGGCCCCCCAGUAAGGCAUGCAAAUAAUUUAAAUCAGCAGGAA------AACACCUGCCGGAC ((((((......))))))...........((((((((-.......(((((((((((...))))))......)))))((...............)).....------....)))))))).. ( -39.36) >DroSim_CAF1 9714 113 - 1 GGCCAAUUGGCUUUGGUUAAAAUUUCUGGCCGGCGGG-GGAAUUUAUGCCGGGGGCAUGGCCCCCCAGUAAGGCAUGCAAAUAAUUUAAAUCAGCAGGAA------AACACCUGCCGGAC ((((((......))))))...........((((((((-.......(((((((((((...))))))......)))))((...............)).....------....)))))))).. ( -39.36) >DroEre_CAF1 12186 119 - 1 GGCCAAUUGGCUUUGGUUAAAAUGUGCGGCCGCCGGGGGGAAUUUAUGCCGGGGGCUUGGC-CCCCAGCAAGGCAUGCAAAUAAUUUAAAUCAGCAGGAAAACGAAAACACCUGCCGGAC ((((....)))).(((((.((((.(((((((.((....))......(((.((((((...))-)))).))).))).))))....)))).)))))(((((............)))))..... ( -42.40) >DroYak_CAF1 7665 119 - 1 GGCCAAUUGGCUUUGGUUAAAAUUUCUGGCCGCCGGG-GGAAUUUAUGCCGGGGGCUUUGCCCCCCAGCAAGGCAUGCAAAUAAUUUAAAUCAGCAGGAAAACGAAAACACCUGCCGGAC (((((.(((((....)))))......))))).(((((-.(((((((((((((((((...))))))......)))))).....)))))...)).(((((............)))))))).. ( -37.00) >consensus GGCCAAUUGGCUUUGGUUAAAAUUUCUGGCCGGCGGG_GGAAUUUAUGCCGGGGGCUUGGCCCCCCAGUAAGGCAUGCAAAUAAUUUAAAUCAGCAGGAA______AACACCUGCCGGAC ((((((......))))))...........((((((((.(......(((((.((((.......)))).....)))))((...............)).............).)))))))).. (-31.80 = -32.20 + 0.40)

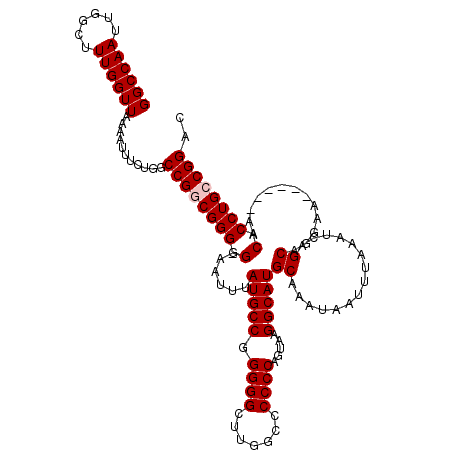
| Location | 8,611,980 – 8,612,099 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.23 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -32.88 |
| Energy contribution | -32.72 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8611980 119 + 20766785 UUGCAUGCCUUACUGGGGUGUCGAGCCCCCGGCAUAAAUUCC-CCCGCCGGCCAGAAAUUUUAACCAAAGCCAAUUGGCCAAGAUAAGAUGUAAUGGUCUUUUAUGGCCCAGUUCGCUGU ..(((.((...((((((((((((......)))))).......-......(((((...((((((.(....(((....)))...).))))))....)))))........))))))..))))) ( -33.50) >DroSec_CAF1 12890 119 + 1 UUGCAUGCCUUACUGGGGGGCCAUGCCCCCGGCAUAAAUUCC-CCCGCCGGCCAGAAAUUUUAACCAAAGCCAAUUGGCCAAGAUAAGAUGUAAUGGUCUUUUAUGGCCCAGUUCGCUGU ..(((.((...((((((((((...))))))(((.........-...)))(((((...............(((....)))......(((((......)))))...)))))))))..))))) ( -37.40) >DroSim_CAF1 9748 119 + 1 UUGCAUGCCUUACUGGGGGGCCAUGCCCCCGGCAUAAAUUCC-CCCGCCGGCCAGAAAUUUUAACCAAAGCCAAUUGGCCAAGAUAAGAUGUAAUGGUCUUUUAUGGCCCAGUUCGCUGU ..(((.((...((((((((((...))))))(((.........-...)))(((((...............(((....)))......(((((......)))))...)))))))))..))))) ( -37.40) >DroEre_CAF1 12226 119 + 1 UUGCAUGCCUUGCUGGGG-GCCAAGCCCCCGGCAUAAAUUCCCCCCGGCGGCCGCACAUUUUAACCAAAGCCAAUUGGCCAAGAUAAGAUGUAAUGGUCUUUUAUGGCCCAGUUCGCUGU .(((..(((.((((((((-((...))))))))))......((....)).))).)))............(((.(((((((((.((.(((((......))))))).))).)))))).))).. ( -43.00) >DroYak_CAF1 7705 119 + 1 UUGCAUGCCUUGCUGGGGGGCAAAGCCCCCGGCAUAAAUUCC-CCCGGCGGCCAGAAAUUUUAACCAAAGCCAAUUGGCCAAGAUAAGAUGUAAUGGUCUUUUAUGGCCCAGUUUGCUGU ......(((..(((((((((....(((...)))......)))-)))))))))................(((.(((((((((.((.(((((......))))))).))).)))))).))).. ( -39.70) >consensus UUGCAUGCCUUACUGGGGGGCCAAGCCCCCGGCAUAAAUUCC_CCCGCCGGCCAGAAAUUUUAACCAAAGCCAAUUGGCCAAGAUAAGAUGUAAUGGUCUUUUAUGGCCCAGUUCGCUGU ..(((.((...((((((((((...))))))((((((((...........(((((...((((((.(....(((....)))...).))))))....))))).))))).)))))))..))))) (-32.88 = -32.72 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:28 2006