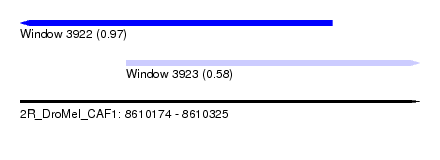
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,610,174 – 8,610,325 |
| Length | 151 |
| Max. P | 0.974622 |

| Location | 8,610,174 – 8,610,292 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.14 |
| Mean single sequence MFE | -30.02 |
| Consensus MFE | -25.62 |
| Energy contribution | -26.02 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974622 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
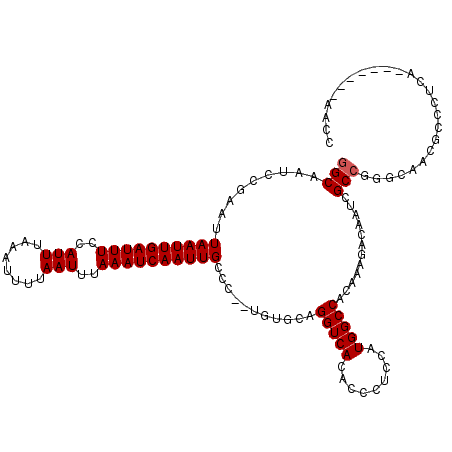
>2R_DroMel_CAF1 8610174 118 - 20766785 GGCCAUGAUGGGUGUGACCUGCACA--GGGCAAUUGAUUUAAAUUAAAAUUUAAAUGGAAAUCAAUUAAUUCGGAUUGCCAGUGAUUUUAAUUGAAAUUCGUAUUCUACGGCCAAAAACU ((((.((....((((.....)))).--...))((.(((((.((((((((((....(((.((((..........)))).)))..)))))))))).))))).)).......))))....... ( -26.60) >DroSec_CAF1 11078 118 - 1 GGCCAUGGUGGGUGUGACCUGCACA--GGGCAAUUGAUUUAAAUUAAAAUUUAAAUGGAAAUCAAUUAAUUCGGAUUGCCAGUGAUUUUAAUUGAAAUUCGUAUUCUACGGCCAAAAACU ((((.(((..((.....))..).))--(((..((.(((((.((((((((((....(((.((((..........)))).)))..)))))))))).))))).))..)))..))))....... ( -28.80) >DroSim_CAF1 7928 118 - 1 GGCCAUGGAGGGUGUGACCUGCACA--GGGCAAUUGAUUUAAAUUAAAAUUUAAAUGGAAAUCAAUUAAUUCGGAUUGCCAGUGAUUUUAAUUGAAAUUCGUAUUCUACGGCCAAAAACU ((((.(((((.((((.....)))).--.....((.(((((.((((((((((....(((.((((..........)))).)))..)))))))))).))))).)).))))).))))....... ( -29.30) >DroEre_CAF1 10382 118 - 1 GGCCAUGGAGGGCGUGACCUGCACA--GGGCAAUUGAUUUAAAUUAAAAUUUAAAUGGAAAUCAAUUAAUUCGGAUUGCCAGUGAUUUUAAUUGAAAUUCGAAUGGUACGGCCAAAAACU ((((.((.(((......))).))..--(..((((((((((..(((........)))..))))))))).(((((((((..((((.......)))).))))))))).)..)))))....... ( -33.10) >DroYak_CAF1 5672 120 - 1 GGCCAUGGAGGGUGUGACCUGCACACGGGGCAAUUGAUUUAAAUUAAAAUUUAAAUGGAAAUCAAUUAAUUCGGAUUGCCAGUGAUUUUAAUUGAAAUUCGUAUUCUACGGCCAAAAACU ((((.(((((.(((((.....)))))(((..(((((((((..(((........)))..)))))))))..)))(((((..((((.......)))).)))))...))))).))))....... ( -32.30) >consensus GGCCAUGGAGGGUGUGACCUGCACA__GGGCAAUUGAUUUAAAUUAAAAUUUAAAUGGAAAUCAAUUAAUUCGGAUUGCCAGUGAUUUUAAUUGAAAUUCGUAUUCUACGGCCAAAAACU ((((.((.(((......))).))........(((((((((..(((........)))..)))))))))....((((((..((((.......)))).))))))........))))....... (-25.62 = -26.02 + 0.40)


| Location | 8,610,214 – 8,610,325 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.66 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -15.60 |
| Energy contribution | -15.80 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8610214 111 + 20766785 GGCAAUCCGAAUUAAUUGAUUUCCAUUUAAAUUUUAAUUUAAAUCAAUUGCCC--UGUGCAGGUCACACCCAUCAUGGCCACAAAGACAAUCGCCAUCCACCGCCCUCA-------AACU (((.....((..((((((((((..(((........)))..)))))))))).((--(....))).........))(((((.............))))).....)))....-------.... ( -17.32) >DroSec_CAF1 11118 111 + 1 GGCAAUCCGAAUUAAUUGAUUUCCAUUUAAAUUUUAAUUUAAAUCAAUUGCCC--UGUGCAGGUCACACCCACCAUGGCCACAAAGACAAGCGCCGGUCAUCGCCCUCA-------AACC (((.........((((((((((..(((........)))..)))))))))).((--.((((..(((....((.....)).......)))..)))).)).....)))....-------.... ( -21.90) >DroSim_CAF1 7968 111 + 1 GGCAAUCCGAAUUAAUUGAUUUCCAUUUAAAUUUUAAUUUAAAUCAAUUGCCC--UGUGCAGGUCACACCCUCCAUGGCCACAAAGACAAGCGCCGGGCAUCGCCCUCA-------AACC (((..........(((((((((..(((........)))..)))))))))((((--.((((..(((....((.....)).......)))..)))).))))...)))....-------.... ( -28.80) >DroEre_CAF1 10422 118 + 1 GGCAAUCCGAAUUAAUUGAUUUCCAUUUAAAUUUUAAUUUAAAUCAAUUGCCC--UGUGCAGGUCACGCCCUCCAUGGCCACAAAGACAAUCGCCUGGCAACUCCCCCAAUCCCCAAACC (((..........(((((((((..(((........)))..)))))))))(((.--((.(.(((......)))))).))).............)))(((........)))........... ( -19.20) >DroYak_CAF1 5712 113 + 1 GGCAAUCCGAAUUAAUUGAUUUCCAUUUAAAUUUUAAUUUAAAUCAAUUGCCCCGUGUGCAGGUCACACCCUCCAUGGCCACAAAGACAAUCGCAUGGCCACUCCCCCA-------AACC (((.((.(((..((((((((((..(((........)))..))))))))))..(((((.(.(((......)))))))))............))).)).))).........-------.... ( -22.30) >consensus GGCAAUCCGAAUUAAUUGAUUUCCAUUUAAAUUUUAAUUUAAAUCAAUUGCCC__UGUGCAGGUCACACCCUCCAUGGCCACAAAGACAAUCGCCGGGCAACGCCCUCA_______AACC (((.........((((((((((..(((........)))..))))))))))...........(((((.........)))))............)))......................... (-15.60 = -15.80 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:22 2006