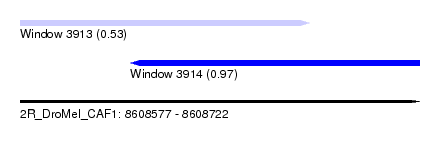
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,608,577 – 8,608,722 |
| Length | 145 |
| Max. P | 0.967294 |

| Location | 8,608,577 – 8,608,682 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.76 |
| Mean single sequence MFE | -28.84 |
| Consensus MFE | -16.72 |
| Energy contribution | -17.52 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.58 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8608577 105 + 20766785 UAUCGAUUGAAGUCGAAAAAUGAGUCAAUCUUCGAUCGGCUUUAAUGGCAACUG-------CAUAGCGG-AAA----UCUGUGCACUCAAUCUGCAUGCAUGCUUCU---AUAUUUGCCA ....((((((((..((........))...))))))))(((..((..((((..((-------(((.((((-(..----..((......)).))))))))))))))..)---).....))). ( -27.90) >DroSec_CAF1 9538 108 + 1 UAUCGAUUGAAGGCGAAAAAUGAGUCAAUCUUCGAUCGGCUUUAAUGGCAACUGACUGCAUCAUAGUGG-GAA----UCUGUGCACUCAAUCUGGU----UGCUUCU---AUAUUUGCCA ....(((((((((.((........))..)))))))))(((..((..(((((((((.((...(((((...-...----.)))))....)).)).)))----))))..)---).....))). ( -27.00) >DroSim_CAF1 6382 109 + 1 UAUCGAUUGAAGGCGAAAAAUGAGUCAAUCUUCGAUCGGCUUUAAUGGCAACUGACUGCAUCAUAGUAGUGAA----UCUGUGCACUCAAUCUGCG----UGCUUCU---AUAUUUGCCA ..(((((((((((.((........))..)))))))))))......((((((.(.(((((......))))).).----.....((((.(.....).)----)))....---....)))))) ( -28.30) >DroEre_CAF1 8974 93 + 1 UAUCGAUUGAAGGCAGAAAA-GAGUCAAUCUUCGAUCGGCUUUACUGGCAACUG-------CAUAG---------------UGCACUCAAUCUGCA----UGCCUCUGCCAUAUUUGCCA ....(((((((((..((...-...))..)))))))))(((.....(((((...(-------((...---------------((((.......))))----)))...))))).....))). ( -28.20) >DroYak_CAF1 4000 105 + 1 ---CGAUUGAAGGCGAAAAAGGAGUCAAUCUUCGAUCGGCUUUAAUGGCAACUG----UGGCACAGUGG-GAAUCUAUCUGUGCACUCAGUCUGCA----UGCUUCU---AUAUUUGCCA ---........((((((...((((((.(((...))).))))))....(((((((----.((((((((((-....))).)))))).).)))).))).----.......---...)))))). ( -32.80) >consensus UAUCGAUUGAAGGCGAAAAAUGAGUCAAUCUUCGAUCGGCUUUAAUGGCAACUG_______CAUAGUGG_GAA____UCUGUGCACUCAAUCUGCA____UGCUUCU___AUAUUUGCCA ...(((((((((..((........))...))))))))).......((((((..........(((((............)))))..........(((....)))...........)))))) (-16.72 = -17.52 + 0.80)

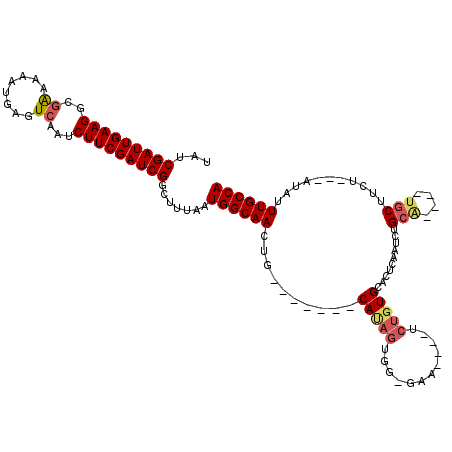
| Location | 8,608,617 – 8,608,722 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.65 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -19.02 |
| Energy contribution | -20.32 |
| Covariance contribution | 1.30 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8608617 105 - 20766785 GGCGGGCUUUUAAUUAAAAGGCCGAAUGGGCCAAAAAUGUUGGCAAAUAU---AGAAGCAUGCAUGCAGAUUGAGUGCACAGA----UUU-CCGCUAUG-------CAGUUGCCAUUAAA ..(((.(((((....))))).)))((((((((((.....)))))......---...(((.(((((((.(((((......))).----..)-).)).)))-------))))).)))))... ( -28.30) >DroSec_CAF1 9578 108 - 1 GGCGUGCUUUUAAUUAAAAGGCCGAAUGGGCCAAAAAUGUUGGCAAAUAU---AGAAGCA----ACCAGAUUGAGUGCACAGA----UUC-CCACUAUGAUGCAGUCAGUUGCCAUUAAA (((.((((((((.......((((.....)))).....((((....)))))---)))))))----....((((((.((((((..----...-......)).)))).)))))))))...... ( -32.40) >DroSim_CAF1 6422 109 - 1 GGCGUGCUUUUAAUUAAAAGGCCGAAUGGGCCAAAAAUGUUGGCAAAUAU---AGAAGCA----CGCAGAUUGAGUGCACAGA----UUCACUACUAUGAUGCAGUCAGUUGCCAUUAAA .(((((((((((.......((((.....)))).....((((....)))))---)))))))----))).((((((.((((((..----..........)).)))).))))))......... ( -36.00) >DroEre_CAF1 9013 94 - 1 GGCGGGCUUUUAAUUAAAAGGCCGAAUGGGCCAAAAAUGCUGGCAAAUAUGGCAGAGGCA----UGCAGAUUGAGUGCA---------------CUAUG-------CAGUUGCCAGUAAA (((.(((((((.....)))))))......))).....(((((((((.....(((.(((((----(.(.....).)))).---------------)).))-------)..))))))))).. ( -33.00) >DroYak_CAF1 4037 108 - 1 GGCGUGCUUUUAAUUAAAAGGCCGAAUGGGCCAAAAAUGUUGGCAAAUAU---AGAAGCA----UGCAGACUGAGUGCACAGAUAGAUUC-CCACUGUGCCA----CAGUUGCCAUUAAA .(((((((((((.......((((.....)))).....((((....)))))---)))))))----))).(((((.(.((((((........-...))))))).----)))))......... ( -36.70) >consensus GGCGUGCUUUUAAUUAAAAGGCCGAAUGGGCCAAAAAUGUUGGCAAAUAU___AGAAGCA____UGCAGAUUGAGUGCACAGA____UUC_CCACUAUG_______CAGUUGCCAUUAAA (((((((.(((((((....((((.....)))).........................((......)).))))))).)))).................(((.....)))...)))...... (-19.02 = -20.32 + 1.30)

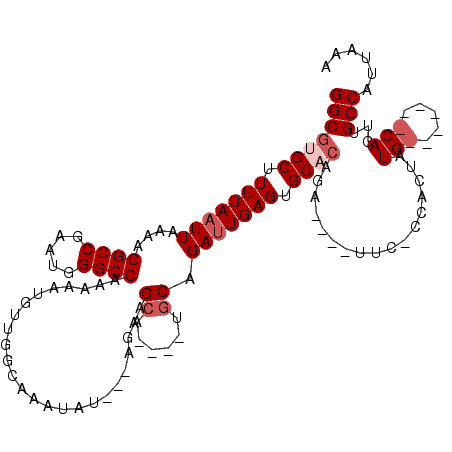
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:14 2006