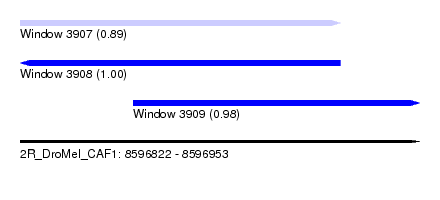
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,596,822 – 8,596,953 |
| Length | 131 |
| Max. P | 0.995360 |

| Location | 8,596,822 – 8,596,927 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 72.55 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -12.81 |
| Energy contribution | -14.40 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8596822 105 + 20766785 CUCUGUCAGCUUGAUUCAUUGUGGAAGUGAAAAUG-AGCCAGCAGGGCGAAAGAAACUGGACGAGGUAAUAGGUC-----CUGGGACAGGUCCUGUCCAACUGACAGAUGG .((((((((.(((.((((((.....))))))....-.....(((((((....)...(((..(.(((........)-----)).)..))).)))))).)))))))))))... ( -36.40) >DroPse_CAF1 30733 92 + 1 CUCUGUCAGCUUGAUUCAUU--CGCAACGAAAGUUUGGGCAGUUG-----------CCGG------AGAUGGCUACGGGGAUGCGACAGGUCCUGCCCUGCUGACAGAUGG .(((((((((..........--...(((....))).((((.((.(-----------(((.------...)))).)).(((((.......))))))))).)))))))))... ( -37.10) >DroSec_CAF1 81158 91 + 1 CUCUGUCAGCUUGAUUCAUUGUGGAAGUGAAAAUG-AGCCAGCCGGGCGAAAGAAACUGGACGAGGUAAUAGGUC-----CUGGGACAGGUCCUGUC-------------- (((.(((.((((..((((((.....))))))...)-)))....(((.(....)...)))))))))...(((((.(-----(((...)))).))))).-------------- ( -27.80) >DroSim_CAF1 73087 105 + 1 CUCUGUCAGCUUGAUUCAUUGUGGAAGUGAAAAUG-AGCCAGCCGGGCGAAAGAAACUGGACGAGGUAAUAGGUC-----CUGGGACAGGUCCUGUCCAACUGACAGAUGG .((((((((.(((.((((((.....))))))....-.(((.(((((.(....)...)))).)..))).(((((.(-----(((...)))).))))).)))))))))))... ( -38.20) >DroYak_CAF1 76099 105 + 1 CUCUGUCAGCUUGAUUCAUUGUGGAAGUGAAAAUG-AGCCAGCAGGGCGAAAGAAACAGGACGAGGUAAUAGGUC-----CUGGGACAGGUCCUGUCCAACUGACAGAUGG .(((((((((((..((((((.....))))))....-.(((.....)))..)))...((((((..........)))-----)))((((((...))))))..))))))))... ( -37.00) >DroPer_CAF1 31012 92 + 1 CUCUGUCAGCUUGAUUCAUU--CGCAACGAAAGUUUGGGCAGUUG-----------CCGG------AGAUGGCUACGGGGAUGCGACAGGUCCUGCCCCGCUGACAGAUGG .(((((((((..........--...(((....))).((((.((.(-----------(((.------...)))).)).(((((.......))))))))).)))))))))... ( -37.00) >consensus CUCUGUCAGCUUGAUUCAUUGUGGAAGUGAAAAUG_AGCCAGCAGGGCGAAAGAAACUGGACGAGGUAAUAGGUC_____CUGGGACAGGUCCUGUCCAACUGACAGAUGG .((((((((.....((((((.....)))))).......((((.....(....)...(((..........)))........))))(((((...)))))...))))))))... (-12.81 = -14.40 + 1.59)

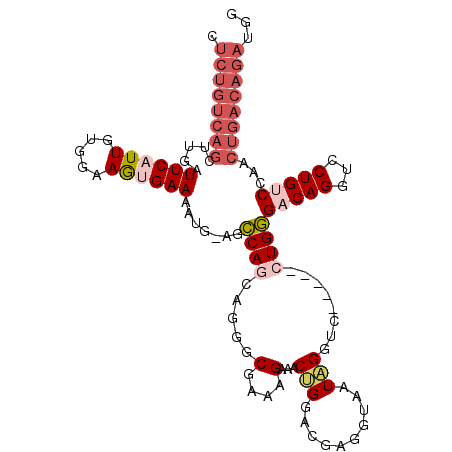
| Location | 8,596,822 – 8,596,927 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 72.55 |
| Mean single sequence MFE | -30.82 |
| Consensus MFE | -14.82 |
| Energy contribution | -16.60 |
| Covariance contribution | 1.78 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.48 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995360 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8596822 105 - 20766785 CCAUCUGUCAGUUGGACAGGACCUGUCCCAG-----GACCUAUUACCUCGUCCAGUUUCUUUCGCCCUGCUGGCU-CAUUUUCACUUCCACAAUGAAUCAAGCUGACAGAG ...(((((((((((((((((....(((....-----)))......))).))))..........(((.....))).-........................)))))))))). ( -30.10) >DroPse_CAF1 30733 92 - 1 CCAUCUGUCAGCAGGGCAGGACCUGUCGCAUCCCCGUAGCCAUCU------CCGG-----------CAACUGCCCAAACUUUCGUUGCG--AAUGAAUCAAGCUGACAGAG ...(((((((((.((((((((....))...........(((....------..))-----------)..)))))).....(((((....--.)))))....))))))))). ( -35.80) >DroSec_CAF1 81158 91 - 1 --------------GACAGGACCUGUCCCAG-----GACCUAUUACCUCGUCCAGUUUCUUUCGCCCGGCUGGCU-CAUUUUCACUUCCACAAUGAAUCAAGCUGACAGAG --------------((.((((.(((..(.((-----(........))).)..))).)))).))...((((((..(-((((...........)))))..).)))))...... ( -20.70) >DroSim_CAF1 73087 105 - 1 CCAUCUGUCAGUUGGACAGGACCUGUCCCAG-----GACCUAUUACCUCGUCCAGUUUCUUUCGCCCGGCUGGCU-CAUUUUCACUUCCACAAUGAAUCAAGCUGACAGAG ...(((((((((((((((((....(((....-----)))......))).))))..........(((.....))).-........................)))))))))). ( -30.10) >DroYak_CAF1 76099 105 - 1 CCAUCUGUCAGUUGGACAGGACCUGUCCCAG-----GACCUAUUACCUCGUCCUGUUUCUUUCGCCCUGCUGGCU-CAUUUUCACUUCCACAAUGAAUCAAGCUGACAGAG ...((((((((((((((((...))))))(((-----(((..........))))))........(((.....))).-........................)))))))))). ( -32.60) >DroPer_CAF1 31012 92 - 1 CCAUCUGUCAGCGGGGCAGGACCUGUCGCAUCCCCGUAGCCAUCU------CCGG-----------CAACUGCCCAAACUUUCGUUGCG--AAUGAAUCAAGCUGACAGAG ...(((((((((.((((((((....))...........(((....------..))-----------)..)))))).....(((((....--.)))))....))))))))). ( -35.60) >consensus CCAUCUGUCAGUUGGACAGGACCUGUCCCAG_____GACCUAUUACCUCGUCCAGUUUCUUUCGCCCAGCUGGCU_CAUUUUCACUUCCACAAUGAAUCAAGCUGACAGAG ...(((((((((..(((((...)))))........................(((((............)))))............................))))))))). (-14.82 = -16.60 + 1.78)

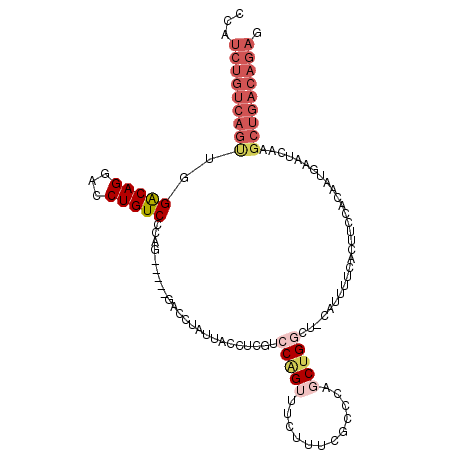
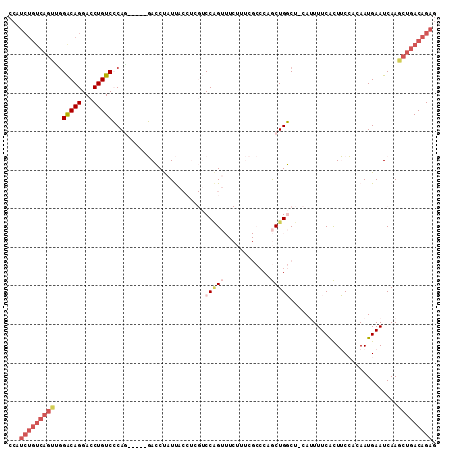
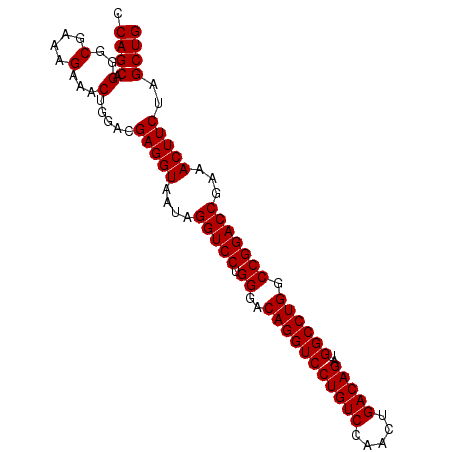
| Location | 8,596,859 – 8,596,953 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 98.58 |
| Mean single sequence MFE | -38.97 |
| Consensus MFE | -37.50 |
| Energy contribution | -37.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8596859 94 + 20766785 CCAGCAGGGCGAAAGAAACUGGACGAGGUAAUAGGUCCUGGGACAGGUCCUGUCCAACUGACAGAUGGCCUGGCCGGACCGAAACUUCUAGCUG .((((((..(....)...))....(((((....(((((.((..(((((((((((.....)))))..)))))).)))))))...)))))..)))) ( -38.80) >DroSim_CAF1 73124 94 + 1 CCAGCCGGGCGAAAGAAACUGGACGAGGUAAUAGGUCCUGGGACAGGUCCUGUCCAACUGACAGAUGGCCUGGCCGGACCGAAACUUCUAGCUG .(((((((.(....)...)))...(((((....(((((.((..(((((((((((.....)))))..)))))).)))))))...)))))..)))) ( -40.00) >DroYak_CAF1 76136 94 + 1 CCAGCAGGGCGAAAGAAACAGGACGAGGUAAUAGGUCCUGGGACAGGUCCUGUCCAACUGACAGAUGGCCUGGCCGGACCGAAACUUCUAGCUG .((((.(..(....)...).....(((((....(((((.((..(((((((((((.....)))))..)))))).)))))))...)))))..)))) ( -38.10) >consensus CCAGCAGGGCGAAAGAAACUGGACGAGGUAAUAGGUCCUGGGACAGGUCCUGUCCAACUGACAGAUGGCCUGGCCGGACCGAAACUUCUAGCUG .((((.(..(....)...).....(((((....(((((.((..(((((((((((.....)))))..)))))).)))))))...)))))..)))) (-37.50 = -37.50 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:09 2006