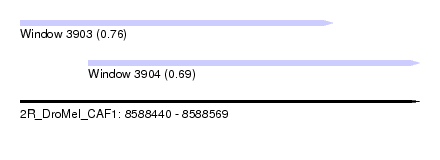
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 8,588,440 – 8,588,569 |
| Length | 129 |
| Max. P | 0.764667 |

| Location | 8,588,440 – 8,588,541 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 75.84 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -14.48 |
| Energy contribution | -15.87 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
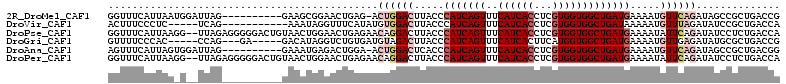
>2R_DroMel_CAF1 8588440 101 + 20766785 GGUUUCAUUAAUGGAUUAG----------GAAGCGGAACUGAG-ACUGGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUGUUCAGAUAGCCGCUGACCG ..................(----------(.(((((..(((..-.((((((.....(((((((..((((((...))))))))))))).....)))))).))))))))..)). ( -31.50) >DroVir_CAF1 20380 96 + 1 ACUUUCCCUC-----UCAG-----------AAAUAGGUUUCAUAUGUGGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUAAAAAUGUUUAGAUAUCCGCUGACCA ..........-----....-----------.....((((..((((.(((((......((((((..((((((...))))))))))))......))))).)))).....)))). ( -21.00) >DroPse_CAF1 22823 110 + 1 GGUUUCAUUAAGG--UUAGAGGGGGACUGUAACUGGAACUGAGAACAGGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUAUUCAGAUAUCCUCUGACCA ...........((--((((((((...(((....(((((((((.....((......)).)))))))))(((((....)))))..............)))...)))))))))). ( -36.20) >DroGri_CAF1 20424 99 + 1 GUUUUCCCAC-----CCAG---GA-----GACAUAGGUCUGUGAUGUAGACUUACCCAUCAGUUUCAUCACUUCAUGGUGGCUGAUGAAAAUGUUGAGAUAUGCGCUGACCG ((((((....-----...)---))-----)))...((((.(((((((.(((.....(((((((..(((((.....)))))))))))).....)))...)))).))).)))). ( -29.50) >DroAna_CAF1 14803 101 + 1 AGUUUCAUUAGUGGAUUAG----------GAAAUGAGACUGGA-ACUGGACUCACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUGUUCAGAUAGCCGCUGACGG ...((((....))))..((----------(..((((((((((.-..(((......)))))))))))))..)))(((((((((((.((((....))))..)))))))).))). ( -33.00) >DroPer_CAF1 23129 110 + 1 GGUUUCAUUAAGG--UUAGAGGGGGACUGUAACUGGAACUGAGAACAGGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUAUUCAGAUAUCCUCUGACCA ...........((--((((((((...(((....(((((((((.....((......)).)))))))))(((((....)))))..............)))...)))))))))). ( -36.20) >consensus GGUUUCAUUAA_G__UUAG__________GAAAUGGAACUGAGAACUGGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUGUUCAGAUAUCCGCUGACCA .............................................((((((.....(((((((..((((((...))))))))))))).....)))))).............. (-14.48 = -15.87 + 1.39)

| Location | 8,588,462 – 8,588,569 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -37.97 |
| Consensus MFE | -21.47 |
| Energy contribution | -22.08 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.689129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 8588462 107 + 20766785 GCGGAACUGAG-ACUGGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUGUUCAGAUAGCCGCUGACCG------------AAGUGGAAAUGGUUAGGCAUAGGAGCAG ((((((((((.-..(((......)))))))))))....(((.((((((((((.((((....))))..)))))))(((((.------------..((....)))))))..)))))).)).. ( -34.50) >DroVir_CAF1 20396 120 + 1 AUAGGUUUCAUAUGUGGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUAAAAAUGUUUAGAUAUCCGCUGACCAAAGGUAGCUGCCAAGUGAUCCAAGAGAUCCAGAGCAGCUG ...((((..((((.(((((......((((((..((((((...))))))))))))......))))).)))).....)))).....(((((((...(.((((.....)))))...))))))) ( -33.60) >DroPse_CAF1 22853 120 + 1 CUGGAACUGAGAACAGGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUAUUCAGAUAUCCUCUGACCAAAGGCGUCCGCCAAGAGGCCAACGAGAGGCAGAGGAGCAG (((...(((((....((.....))(((((((..((((((...)))))))))))))......)))))...(((((((.((.....(((..(((....)))..)))...))))))))).))) ( -40.80) >DroEre_CAF1 54711 107 + 1 GCGGAAAUGAG-ACUGGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUGUUCAGAUAGCCGCUGACCG------------AGGUGGACAUGGUUAGGCAUAGGAGCAG ((.....((((-(((((.........)))))))))((((((((.((((((((.((((....))))..))))))))...))------------))))))..................)).. ( -37.10) >DroAna_CAF1 14825 119 + 1 AUGAGACUGGA-ACUGGACUCACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUGUUCAGAUAGCCGCUGACGGAGGGCGUCCGCCAAGUGGAAAUGGUCAGGAAGAGGAGCCA ((((((((((.-..(((......)))))))))))))..(..(((((((((((.((((....))))..)))))))).)))..)(((.(((((((........))))..)))......))). ( -41.00) >DroPer_CAF1 23159 120 + 1 CUGGAACUGAGAACAGGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUAUUCAGAUAUCCUCUGACCAAAGGCGUCCGCCAAGAGGCCAACGAGAGGCAGAGGAGCAG (((...(((((....((.....))(((((((..((((((...)))))))))))))......)))))...(((((((.((.....(((..(((....)))..)))...))))))))).))) ( -40.80) >consensus AUGGAACUGAG_ACUGGACUUACCCAUCAGUUUCAUCACCUCGUGGUGGCUGAUGAAAAUGUUCAGAUAGCCGCUGACCAAAGGCGUCCGCCAAGUGGACAAGGAGAGGCAGAGGAGCAG .............((((((.....(((((((..((((((...))))))))))))).....))))))....(((((.......(((....))).)))))...................... (-21.47 = -22.08 + 0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:05 2006